Strain belonging to an emerging, virulent sublineage of ST131 Escherichia coli isolated in fresh spinach, suggesting that ST131 may be transmissible through agricultural products
- PMID: 37876872
- PMCID: PMC10591226
- DOI: 10.3389/fcimb.2023.1237725
Strain belonging to an emerging, virulent sublineage of ST131 Escherichia coli isolated in fresh spinach, suggesting that ST131 may be transmissible through agricultural products
Abstract
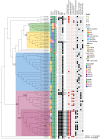
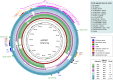
Food contamination with pathogenic Escherichia coli can cause severe disease. Here, we report the isolation of a multidrug resistant strain (A23EC) from fresh spinach. A23EC belongs to subclade C2 of ST131, a virulent clone of Extraintestinal Pathogenic E. coli (ExPEC). Most A23EC virulence factors are concentrated in three pathogenicity islands. These include PapGII, a fimbrial tip adhesin linked to increased virulence, and CsgA and CsgB, two adhesins known to facilitate spinach leaf colonization. A23EC also bears TnMB1860, a chromosomally-integrated transposon with the demonstrated potential to facilitate the evolution of carbapenem resistance among non-carbapenemase-producing enterobacterales. This transposon consists of two IS26-bound modular translocatable units (TUs). The first TU carries aac(6')-lb-cr, bla OXA-1, ΔcatB3, aac(3)-lle, and tmrB, and the second one harbors bla CXT-M-15. A23EC also bears a self-transmissible plasmid that can mediate conjugation at 20°C and that has a mosaic IncF [F(31,36):A(4,20):B1] and Col156 origin of replication. Comparing A23EC to 86 additional complete ST131 sequences, A23EC forms a monophyletic cluster with 17 other strains that share the following four genomic traits: (1) virotype E (papGII+); (2) presence of a PAI II536-like pathogenicity island with an additional cnf1 gene; (3) presence of chromosomal TnMB1860; and (4) frequent presence of an F(31,36):A(4,20):B1 plasmid. Sequences belonging to this cluster (which we named "C2b sublineage") are highly enriched in septicemia samples and their associated genetic markers align with recent reports of an emerging, virulent sublineage of the C2 subclade, suggesting significant pathogenic potential. This is the first report of a ST131 strain belonging to subclade C2 contaminating green leafy vegetables. The detection of this uropathogenic clone in fresh food is alarming. This work suggests that ST131 continues to evolve, gaining selective advantages and new routes of transmission. This highlights the pressing need for rigorous epidemiological surveillance of ExPEC in vegetables with One Health perspective.
Keywords: ExPEC; ST131; conjugative transfer; food safety; mobile genetic elements; virulence.
Copyright © 2023 Balbuena-Alonso, Camps, Cortés-Cortés, Carreón-León, Lozano-Zarain and Rocha-Gracia.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


Similar articles
-
Arrangements of Mobile Genetic Elements among Virotype E Subpopulation of Escherichia coli Sequence Type 131 Strains with High Antimicrobial Resistance and Virulence Gene Content.mSphere. 2021 Aug 25;6(4):e0055021. doi: 10.1128/mSphere.00550-21. Epub 2021 Aug 25. mSphere. 2021. PMID: 34431692 Free PMC article.
-
Comparative Genomic Analysis of Globally Dominant ST131 Clone with Other Epidemiologically Successful Extraintestinal Pathogenic Escherichia coli (ExPEC) Lineages.mBio. 2017 Oct 24;8(5):e01596-17. doi: 10.1128/mBio.01596-17. mBio. 2017. PMID: 29066550 Free PMC article.
-
Virulence and resistance properties of E. coli isolated from urine samples of hospitalized patients in Rio de Janeiro, Brazil - The role of mobile genetic elements.Int J Med Microbiol. 2020 Dec;310(8):151453. doi: 10.1016/j.ijmm.2020.151453. Epub 2020 Sep 28. Int J Med Microbiol. 2020. PMID: 33045580
-
The Significance of Epidemic Plasmids in the Success of Multidrug-Resistant Drug Pandemic Extraintestinal Pathogenic Escherichia coli.Infect Dis Ther. 2023 Apr;12(4):1029-1041. doi: 10.1007/s40121-023-00791-4. Epub 2023 Mar 22. Infect Dis Ther. 2023. PMID: 36947392 Free PMC article. Review.
-
Escherichia coli ST131: a multidrug-resistant clone primed for global domination.F1000Res. 2017 Feb 28;6:F1000 Faculty Rev-195. doi: 10.12688/f1000research.10609.1. eCollection 2017. F1000Res. 2017. PMID: 28344773 Free PMC article. Review.
Cited by
-
A Novel Bifidobacterium longum Subsp. longum T1 Strain from Cow's Milk: Homeostatic and Antibacterial Activity against ESBL-Producing Escherichia coli.Antibiotics (Basel). 2024 Sep 27;13(10):924. doi: 10.3390/antibiotics13100924. Antibiotics (Basel). 2024. PMID: 39452191 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

