The nuclear pore Y-complex functions as a platform for transcriptional regulation of FLOWERING LOCUS C in Arabidopsis
- PMID: 37877462
- PMCID: PMC10827314
- DOI: 10.1093/plcell/koad271
The nuclear pore Y-complex functions as a platform for transcriptional regulation of FLOWERING LOCUS C in Arabidopsis
Abstract
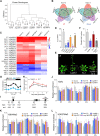
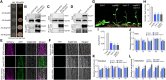
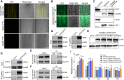
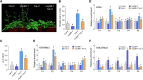
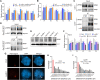
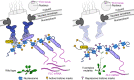
The nuclear pore complex (NPC) has multiple functions beyond the nucleo-cytoplasmic transport of large molecules. Subnuclear compartmentalization of chromatin is critical for gene expression in animals and yeast. However, the mechanism by which the NPC regulates gene expression is poorly understood in plants. Here we report that the Y-complex (Nup107-160 complex, a subcomplex of the NPC) self-maintains its nucleoporin homeostasis and modulates FLOWERING LOCUS C (FLC) transcription via changing histone modifications at this locus. We show that Y-complex nucleoporins are intimately associated with FLC chromatin through their interactions with histone H2A at the nuclear membrane. Fluorescence in situ hybridization assays revealed that Nup96, a Y-complex nucleoporin, enhances FLC positioning at the nuclear periphery. Nup96 interacted with HISTONE DEACETYLASE 6 (HDA6), a key repressor of FLC expression via histone modification, at the nuclear membrane to attenuate HDA6-catalyzed deposition at the FLC locus and change histone modifications. Moreover, we demonstrate that Y-complex nucleoporins interact with RNA polymerase II to increase its occupancy at the FLC locus, facilitating transcription. Collectively, our findings identify an attractive mechanism for the Y-complex in regulating FLC expression via tethering the locus at the nuclear periphery and altering its histone modification.
© The Author(s) 2023. Published by Oxford University Press on behalf of American Society of Plant Biologists. All rights reserved. For permissions, please e-mail: journals.permissions@oup.com.
Conflict of interest statement
Conflict of interest statement. The authors declare no competing interests.
Figures


Similar articles
-
HISTONE DEACETYLASE6 interacts with FLOWERING LOCUS D and regulates flowering in Arabidopsis.Plant Physiol. 2011 May;156(1):173-84. doi: 10.1104/pp.111.174417. Epub 2011 Mar 10. Plant Physiol. 2011. PMID: 21398257 Free PMC article.
-
MSI1 and HDA6 function interdependently to control flowering time via chromatin modifications.Plant J. 2022 Feb;109(4):831-843. doi: 10.1111/tpj.15596. Epub 2021 Nov 30. Plant J. 2022. PMID: 34807487
-
Arabidopsis inositol polyphosphate multikinase delays flowering time through mediating transcriptional activation of FLOWERING LOCUS C.J Exp Bot. 2017 Dec 16;68(21-22):5787-5800. doi: 10.1093/jxb/erx397. J Exp Bot. 2017. PMID: 29161428 Free PMC article.
-
Role of chromatin modification in flowering-time control.Trends Plant Sci. 2005 Jan;10(1):30-5. doi: 10.1016/j.tplants.2004.11.003. Trends Plant Sci. 2005. PMID: 15642521 Review.
-
Post-translational modifications of FLOWERING LOCUS C modulate its activity.J Exp Bot. 2017 Jan 1;68(3):383-389. doi: 10.1093/jxb/erw431. J Exp Bot. 2017. PMID: 28204510 Review.
Cited by
-
Exportin-1 functions as an adaptor for transcription factor-mediated docking of chromatin at the nuclear pore complex.bioRxiv [Preprint]. 2024 May 13:2024.05.09.593355. doi: 10.1101/2024.05.09.593355. bioRxiv. 2024. Update in: Mol Cell. 2025 Mar 20;85(6):1101-1116.e8. doi: 10.1016/j.molcel.2025.02.013. PMID: 38798450 Free PMC article. Updated. Preprint.
-
Exportin-1 functions as an adaptor for transcription factor-mediated docking of chromatin at the nuclear pore complex.Mol Cell. 2025 Mar 20;85(6):1101-1116.e8. doi: 10.1016/j.molcel.2025.02.013. Epub 2025 Mar 10. Mol Cell. 2025. PMID: 40068679
-
Location, location, location: Nuclear pore complexes tether flowering loci to nuclear envelope to boost gene expression.Plant Cell. 2024 Jan 30;36(2):217-218. doi: 10.1093/plcell/koad282. Plant Cell. 2024. PMID: 37943685 Free PMC article. No abstract available.
-
Human Cytomegalovirus UL78 is a Nuclear-Localized GPCR Necessary for Efficient Reactivation from Latent Infection in CD34+ Hematopoietic Progenitor Cells.bioRxiv [Preprint]. 2025 Jun 2:2025.06.02.657350. doi: 10.1101/2025.06.02.657350. bioRxiv. 2025. PMID: 40501644 Free PMC article. Preprint.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

