Genotypic Evolution of Klebsiella pneumoniae Sequence Type 512 during Ceftazidime/Avibactam, Meropenem/Vaborbactam, and Cefiderocol Treatment, Italy
- PMID: 37877547
- PMCID: PMC10617348
- DOI: 10.3201/eid2911.230921
Genotypic Evolution of Klebsiella pneumoniae Sequence Type 512 during Ceftazidime/Avibactam, Meropenem/Vaborbactam, and Cefiderocol Treatment, Italy
Abstract
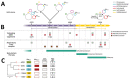
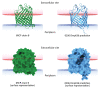
In February 2022, a critically ill patient colonized with a carbapenem-resistant K. pneumoniae producing KPC-3 and VIM-1 carbapenemases was hospitalized for SARS-CoV-2 in the intensive care unit of Policlinico Umberto I hospital in Rome, Italy. During 95 days of hospitalization, ceftazidime/avibactam, meropenem/vaborbactam, and cefiderocol were administered consecutively to treat 3 respiratory tract infections sustained by different bacterial agents. Those therapies altered the resistome of K. pneumoniae sequence type 512 colonizing or infecting the patient during the hospitalization period. In vivo evolution of the K. pneumoniae sequence type 512 resistome occurred through plasmid loss, outer membrane porin alteration, and a nonsense mutation in the cirA siderophore gene, resulting in high levels of cefiderocol resistance. Cross-selection can occur between K. pneumoniae and treatments prescribed for other infective agents. K. pneumoniae can stably colonize a patient, and antimicrobial-selective pressure can promote progressive K. pneumoniae resistome evolution, indicating a substantial public health threat.
Keywords: Acinetobacter baumannii; CirA; Italy; KPC-154; KPC-3; KPC-31; Klebsiella pneumoniae; Providencia stuartii; ST512; VIM-1; antimicrobial resistance; avibactam; bacteria; carbapenemase; carbapenemase co-production; cefiderocol; ceftazidime; colistin resistance; fosfomycin resistance; inhibitor-resistant variants; meropenem; outer membrane porin disruption; respiratory infections; sequence type 512; siderophore disruption; vaborbactam.
Figures

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

