Ac4C Enhances the Translation Efficiency of Vegfa mRNA and Mediates Central Sensitization in Spinal Dorsal Horn in Neuropathic Pain
- PMID: 37877615
- PMCID: PMC10724395
- DOI: 10.1002/advs.202303113
Ac4C Enhances the Translation Efficiency of Vegfa mRNA and Mediates Central Sensitization in Spinal Dorsal Horn in Neuropathic Pain
Abstract
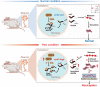
N4-Acetylcytidine (ac4C), a highly conserved post-transcriptional machinery with extensive existence for RNA modification, plays versatile roles in various cellular processes and functions. However, the molecular mechanism by which ac4C modification mediates neuropathic pain remains elusive. Here, it is found that the enhanced ac4C modification promotes the recruitment of polysome in Vegfa mRNA and strengthens the translation efficiency following SNI. Nerve injury increases the expression of NAT10 and the interaction between NAT10 and Vegfa mRNA in the dorsal horn neurons, and the gain and loss of NAT10 function further confirm that NAT10 is involved in the ac4C modification in Vegfa mRNA and pain behavior. Moreover, the ac4C-mediated VEGFA upregulation contributes to the central sensitivity and neuropathic pain induced by SNI or AAV-hSyn-NAT10. Finally, SNI promotes the binding of HNRNPK in Vegfa mRNA and subsequently recruits the NAT10. The enhanced interaction between HNRNPK and NAT10 contributes to the ac4C modification of Vegfa mRNA and neuropathic pain. These findings suggest that the enhanced interaction between HNRNPK and Vegfa mRNA upregulates the ac4C level by recruiting NAT10 and contributes to the central sensitivity and neuropathic pain following SNI. Blocking this cascade may be a novel therapeutic approach in patients with neuropathic pain.
Keywords: NAT10; VEGFA; ac4C modification; neuropathic pain; spinal dorsal horn; translation efficiency.
© 2023 The Authors. Advanced Science published by Wiley-VCH GmbH.
Conflict of interest statement
The authors declare no conflict of interest.
Figures






References
-
- O'connor A. B., Pharmacoeconomics 2009, 27, 95. - PubMed
-
- a) Haute L. V., Hendrick A. G., D'souza A. R., Powell C. A., Rebelo‐Guiomar P., Harbour M. E., Ding S., Fearnley I. M., Andrews B., Minczuk M., Nucleic Acids Res. 2019, 47, 10267; - PMC - PubMed
- b) Liu S., Zhang Y., Qiu L., Zhang S., Meng Y., Huang C., Chen Z., Zhang B., Han J., Front. Cell Dev. Biol. 2022, 10, 861000. - PMC - PubMed
-
- Dominissini D., Moshitch‐Moshkovitz S., Schwartz S., Salmon‐Divon M., Ungar L., Osenberg S., Cesarkas K., Jacob‐Hirsch J., Amariglio N., Kupiec M., Sorek R., Rechavi G., Nature 2012, 485, 201. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- 2021YFA1300201/National Key R&D Program of China
- 20226060004/Guangzhou Science and Technology Plan Project
- 202201010988/Guangzhou Science and Technology Plan Project
- 2022M723652/China Postdoctoral Science Foundation
- 2019B151502010/Basic and Applied Basic Research Foundation of Guangdong Province
- 2022A1515012259/Basic and Applied Basic Research Foundation of Guangdong Province
- 2023A1515030020/Basic and Applied Basic Research Foundation of Guangdong Province
- 31970936/National Natural Science Foundation of China
- 82201377/National Natural Science Foundation of China
- 82201373/National Natural Science Foundation of China
- 81870878/National Natural Science Foundation of China
LinkOut - more resources
Full Text Sources
Miscellaneous
