Rates and Classification of Variants of Uncertain Significance in Hereditary Disease Genetic Testing
- PMID: 37878314
- PMCID: PMC10600581
- DOI: 10.1001/jamanetworkopen.2023.39571
Rates and Classification of Variants of Uncertain Significance in Hereditary Disease Genetic Testing
Abstract
Importance: Variants of uncertain significance (VUSs) are rampant in clinical genetic testing, frustrating clinicians, patients, and laboratories because the uncertainty hinders diagnoses and clinical management. A comprehensive assessment of VUSs across many disease genes is needed to guide efforts to reduce uncertainty.
Objective: To describe the sources, gene distribution, and population-level attributes of VUSs and to evaluate the impact of the different types of evidence used to reclassify them.
Design, setting, and participants: This cohort study used germline DNA variant data from individuals referred by clinicians for diagnostic genetic testing for hereditary disorders. Participants included individuals for whom gene panel testing was conducted between September 9, 2014, and September 7, 2022. Data were analyzed from September 1, 2022, to April 1, 2023.
Main outcomes and measures: The outcomes of interest were VUS rates (stratified by age; clinician-reported race, ethnicity, and ancestry groups; types of gene panels; and variant attributes), percentage of VUSs reclassified as benign or likely benign vs pathogenic or likely pathogenic, and enrichment of evidence types used for reclassifying VUSs.
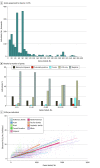
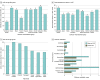
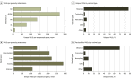
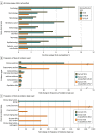
Results: The study cohort included 1 689 845 individuals ranging in age from 0 to 89 years at time of testing (median age, 50 years), with 1 203 210 (71.2%) female individuals. There were 39 150 Ashkenazi Jewish individuals (2.3%), 64 730 Asian individuals (3.8%), 126 739 Black individuals (7.5%), 5539 French Canadian individuals (0.3%), 169 714 Hispanic individuals (10.0%), 5058 Native American individuals (0.3%), 2696 Pacific Islander individuals (0.2%), 4842 Sephardic Jewish individuals (0.3%), and 974 383 White individuals (57.7%). Among all individuals tested, 692 227 (41.0%) had at least 1 VUS and 535 385 (31.7%) had only VUS results. The number of VUSs per individual increased as more genes were tested, and most VUSs were missense changes (86.6%). More VUSs were observed per sequenced gene in individuals who were not from a European White population, in middle-aged and older adults, and in individuals who underwent testing for disorders with incomplete penetrance. Of 37 699 unique VUSs that were reclassified, 30 239 (80.2%) were ultimately categorized as benign or likely benign. A mean (SD) of 30.7 (20.0) months elapsed for VUSs to be reclassified to benign or likely benign, and a mean (SD) of 22.4 (18.9) months elapsed for VUSs to be reclassified to pathogenic or likely pathogenic. Clinical evidence contributed most to reclassification.
Conclusions and relevance: This cohort study of approximately 1.6 million individuals highlighted the need for better methods for interpreting missense variants, increased availability of clinical and experimental evidence for variant classification, and more diverse representation of race, ethnicity, and ancestry groups in genomic databases. Data from this study could provide a sound basis for understanding the sources and resolution of VUSs and navigating appropriate next steps in patient care.
Conflict of interest statement
Figures
References
-
- Richards S, Aziz N, Bale S, et al. ; ACMG Laboratory Quality Assurance Committee . Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17(5):405-424. doi:10.1038/gim.2015.30 - DOI - PMC - PubMed
-
- National Library of Medicine . ClinVar. Accessed September 19, 2023. https://www.ncbi.nlm.nih.gov/clinvar/
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

