The SIB Swiss Institute of Bioinformatics Semantic Web of data
- PMID: 37878411
- PMCID: PMC10767860
- DOI: 10.1093/nar/gkad902
The SIB Swiss Institute of Bioinformatics Semantic Web of data
Abstract
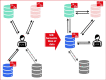
The SIB Swiss Institute of Bioinformatics (https://www.sib.swiss/) is a federation of bioinformatics research and service groups. The international life science community in academia and industry has been accessing the freely available databases provided by SIB since its inception in 1998. In this paper we present the 11 databases which currently offer semantically enriched data in accordance with the FAIR principles (Findable, Accessible, Interoperable, Reusable), as well as the Swiss Personalized Health Network initiative (SPHN) which also employs this enrichment. The semantic enrichment facilitates the manipulation of large data sets from public databases and private data sets. Examples are provided to illustrate that the data from the SIB databases can not only be queried using precise criteria individually, but also across multiple databases, including a variety of non-SIB databases. Data manipulation, be it exploration, extraction, annotation, combination, and publication, is possible using the SPARQL query language. Providing documentation, tutorials and sample queries makes it easier to navigate this web of semantic data. Through this paper, the reader will discover how the existing SIB knowledge graphs can be leveraged to tackle the complex biological or clinical questions that are being addressed today.
© The Author(s) 2023. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures


References
-
- Holmes D.E. 1. The data explosion. Big Data: A Very Short Introduction. 2017; Oxford University Press; 1–13.
-
- Szklarczyk D., Gable A.L., Nastou K.C., Lyon D., Kirsch R., Pyysalo S., Doncheva N.T., Legeay M., Fang T., Bork P.et al. .. The STRING database in 2021: customizable protein–protein networks, and functional characterization of user-uploaded gene/measurement sets. Nucleic Acids Res. 2021; 49:D605–D612. - PMC - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

