HIV-1 subtype diversity and immuno-virological outcomes among adolescents failing antiretroviral therapy in Cameroon: A cohort study
- PMID: 37878637
- PMCID: PMC10599502
- DOI: 10.1371/journal.pone.0293326
HIV-1 subtype diversity and immuno-virological outcomes among adolescents failing antiretroviral therapy in Cameroon: A cohort study
Abstract
Objective: We sought to evaluate the variability of HIV-1 and its effect on immuno-virological response among adolescents living with perinatally acquired HIV (APHI).
Methods: A cohort study was conducted from 2018-2020 among 311 APHI receiving antiretroviral therapy (ART) in Cameroon. Sequencing of protease and reverse transcriptase regions was performed for participants experiencing virological failure, VF, (Plasma viral load, PVL ≥ 1000 RNA copies/ml). HIV-1 subtypes were inferred by phylogeny; immuno-virological responses were monitored at 3-time points (T1-T3). Cox regression modeling was used to estimate adjusted hazard ratios (aHRs) of progression to: CD4 < 250, and PVL > 5log10, adjusted for acquired drug resistance, gender, ART line, adherence, and duration on treatment; p < 0.05 was considered statistically significant.
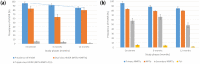
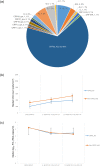
Results: Of the 141 participants in VF enrolled, the male-female ratio was 1:1; mean age was 15 (±3) years; and median [IQR] duration on ART was 51 [46-60] months. In all phases, 17 viral clades were found with a predominant CRF02_AG (58.2%, 59.4%, and 58.3%). From T1-T3 respectively, there was an increasing CD4 count (213 [154-313], 366 [309-469], and 438 [364-569] cells/mm3) and decline log10 PVL (5.23, 4.43, and 4.43), similar across subtypes. Among participants with CRF02_AG infection, duration of treatment was significantly associated with both rates of progression to CD4 < 250, and PVL > 5log10, aHR = 0.02 (0.001-0.52), and aHR = 0.05 (0.01-0.47) respectively. Moreover, four potential new HIV-1 recombinants were identified (CRF02_AG/02D, CRF02_AG/02A1F2, D/CRF02_AG, and AF2/CRF02_AG), indicating a wide viral diversity.
Conclusion: Among APHI in settings like Cameroon, there is a wide genetic diversity of HIV-1, driven by CRF02_AG and with potential novel clades due to ongoing recombination events. Duration of treatment significantly reduces the risk of disease progression.
Copyright: © 2023 Togna Pabo et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

Similar articles
-
Archiving of mutations in HIV-1 cellular reservoirs among vertically infected adolescents is contingent with clinical stages and plasma viral load: Evidence from the EDCTP-READY study.HIV Med. 2022 Jul;23(6):629-638. doi: 10.1111/hiv.13220. Epub 2021 Dec 23. HIV Med. 2022. PMID: 34951111
-
Pre-treatment drug resistance and HIV-1 genetic diversity in the rural and urban settings of Northwest-Cameroon.PLoS One. 2020 Jul 21;15(7):e0235958. doi: 10.1371/journal.pone.0235958. eCollection 2020. PLoS One. 2020. PMID: 32692778 Free PMC article.
-
Rates of Viral Non-Suppression and Acquired HIV-1 Drug Resistance Emergence among Children during the Sociopolitical Crisis in the Northwest Region of Cameroon: A Call for Improved Monitoring Strategies.Curr HIV Res. 2024;22(5):336-348. doi: 10.2174/011570162X319028240830064946. Curr HIV Res. 2024. PMID: 39377407
-
Genotypic resistance testing improves antiretroviral treatment outcomes in a cohort of adolescents in Cameroon: Implications in the dolutegravir-era.J Public Health Afr. 2023 Oct 1;14(10):2612. doi: 10.4081/jphia.2023.2612. eCollection 2023 Oct 31. J Public Health Afr. 2023. PMID: 38020274 Free PMC article.
-
Virological response, HIV-1 drug resistance mutations and genetic diversity among patients on first-line antiretroviral therapy in N'Djamena, Chad: findings from a cross-sectional study.BMC Res Notes. 2017 Nov 10;10(1):589. doi: 10.1186/s13104-017-2893-1. BMC Res Notes. 2017. PMID: 29126456 Free PMC article.
Cited by
-
HIV-1 cross-resistance to second-generation non-nucleoside reverse transcriptase inhibitors among individuals failing antiretroviral therapy in Cameroon: implications for the use of long-acting treatment regimens in low- and middle-income countries.JAC Antimicrob Resist. 2025 Apr 28;7(2):dlaf059. doi: 10.1093/jacamr/dlaf059. eCollection 2025 Apr. JAC Antimicrob Resist. 2025. PMID: 40297528 Free PMC article.
-
Empowering adolescents living with perinatally-acquired HIV: tailored CD4+ count assessment for optimized care, the EDCTP READY-study.Front Med (Lausanne). 2024 Sep 20;11:1457501. doi: 10.3389/fmed.2024.1457501. eCollection 2024. Front Med (Lausanne). 2024. PMID: 39371336 Free PMC article.
References
-
- Banin AN, Tuen M, Bimela JS, Tongo M, Zappile P, Khodadadi‐Jamayran A, et al.. Near full genome characterization of HIV‐1 unique recombinant forms in Cameroon reveals dominant CRF02_AG and F2 recombination patterns. J Intern AIDS Soc. 2019. Jul;22(7):e25362. doi: 10.1002/jia2.25362 - DOI - PMC - PubMed
-
- Kiguoya MW, Mann JK, Chopera D, Gounder K, Lee GQ, Hunt PW, et al.. Subtype-Specific Differences in Gag-Protease-Driven Replication Capacity Are Consistent with Intersubtype Differences in HIV-1 Disease Progression. J Virol. 2017. Jun 9;91(13):e00253–17. doi: 10.1128/JVI.00253-17 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials

