Genetic vulnerability to Crohn's disease reveals a spatially resolved epithelial restitution program
- PMID: 37878672
- PMCID: PMC10798370
- DOI: 10.1126/scitranslmed.adg5252
Genetic vulnerability to Crohn's disease reveals a spatially resolved epithelial restitution program
Abstract
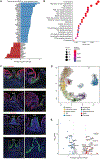
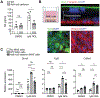
Effective tissue repair requires coordinated intercellular communication to sense damage, remodel the tissue, and restore function. Here, we dissected the healing response in the intestinal mucosa by mapping intercellular communication at single-cell resolution and integrating with spatial transcriptomics. We demonstrated that a risk variant for Crohn's disease, hepatocyte growth factor activator (HGFAC) Arg509His (R509H), disrupted a damage-sensing pathway connecting the coagulation cascade to growth factors that drive the differentiation of wound-associated epithelial (WAE) cells and production of a localized retinoic acid (RA) gradient to promote fibroblast-mediated tissue remodeling. Specifically, we showed that HGFAC R509H was activated by thrombin protease activity but exhibited impaired proteolytic activation of the growth factor macrophage-stimulating protein (MSP). In Hgfac R509H mice, reduced MSP activation in response to wounding of the colon resulted in impaired WAE cell induction and delayed healing. Through integration of single-cell transcriptomics and spatial transcriptomics, we demonstrated that WAE cells generated RA in a spatially restricted region of the wound site and that mucosal fibroblasts responded to this signal by producing extracellular matrix and growth factors. We further dissected this WAE cell-fibroblast signaling circuit in vitro using a genetically tractable organoid coculture model. Collectively, these studies exploited a genetic perturbation associated with human disease to disrupt a fundamental biological process and then reconstructed a spatially resolved mechanistic model of tissue healing.
Conflict of interest statement
Competing interests
R.J.X. is a co-founder of Celsius Therapeutics and Jnana Therapeutics, Scientific Advisory Board member at Nestlé, and Board Director at MoonLake Immunotherapeutics. M.J.D. is a scientific founder of Maze Therapeutics. These organizations had no roles in this study. All other authors declare no competing interests.
Figures





References
-
- Singer AJ, Clark RA, Cutaneous wound healing. N. Engl. J. Med 341, 738–746 (1999). - PubMed
-
- Gurtner GC, Werner S, Barrandon Y, Longaker MT, Wound repair and regeneration. Nature. 453, 314–321 (2008). - PubMed
-
- Harrison OJ, Linehan JL, Shih H-Y, Bouladoux N, Han S-J, Smelkinson M, Sen SK, Byrd AL, Enamorado M, Yao C, Tamoutounour S, Van Laethem F, Hurabielle C, Collins N, Paun A, Salcedo R, O’Shea JJ, Belkaid Y, Commensal-specific T cell plasticity promotes rapid tissue adaptation to injury. Science. 363 (2019), doi:10.1126/science.aat6280. - DOI - PMC - PubMed
-
- Jain U, Ver Heul AM, Xiong S, Gregory MH, Demers EG, Kern JT, Lai C-W, Muegge BD, Barisas DAG, Leal-Ekman JS, Deepak P, Ciorba MA, Liu T-C, Hogan DA, Debbas P, Braun J, McGovern DPB, Underhill DM, Stappenbeck TS, Debaryomyces is enriched in Crohn’s disease intestinal tissue and impairs healing in mice. Science. 371, 1154–1159 (2021). - PMC - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

