TMPRSS2 is a functional receptor for human coronavirus HKU1
- PMID: 37879362
- PMCID: PMC11331971
- DOI: 10.1038/s41586-023-06761-7
TMPRSS2 is a functional receptor for human coronavirus HKU1
Abstract
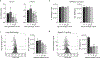
Four endemic seasonal human coronaviruses causing common colds circulate worldwide: HKU1, 229E, NL63 and OC43 (ref. 1). After binding to cellular receptors, coronavirus spike proteins are primed for fusion by transmembrane serine protease 2 (TMPRSS2) or endosomal cathepsins2-9. NL63 uses angiotensin-converting enzyme 2 as a receptor10, whereas 229E uses human aminopeptidase-N11. HKU1 and OC43 spikes bind cells through 9-O-acetylated sialic acid, but their protein receptors remain unknown12. Here we show that TMPRSS2 is a functional receptor for HKU1. TMPRSS2 triggers HKU1 spike-mediated cell-cell fusion and pseudovirus infection. Catalytically inactive TMPRSS2 mutants do not cleave HKU1 spike but allow pseudovirus infection. Furthermore, TMPRSS2 binds with high affinity to the HKU1 receptor binding domain (Kd 334 and 137 nM for HKU1A and HKU1B genotypes) but not to SARS-CoV-2. Conserved amino acids in the HKU1 receptor binding domain are essential for binding to TMPRSS2 and pseudovirus infection. Newly designed anti-TMPRSS2 nanobodies potently inhibit HKU1 spike attachment to TMPRSS2, fusion and pseudovirus infection. The nanobodies also reduce infection of primary human bronchial cells by an authentic HKU1 virus. Our findings illustrate the various evolution strategies of coronaviruses, which use TMPRSS2 to either directly bind to target cells or prime their spike for membrane fusion and entry.
© 2023. The Author(s), under exclusive licence to Springer Nature Limited.
Conflict of interest statement
Conflict of interest
NS, JB, OS, IF, EB, FAR, and PL have a provisional patent on anti-TMPRSS2 nanobodies.
Figures
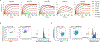









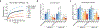

References
Additional references
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

