Mutant p53 sustains serine-glycine synthesis and essential amino acids intake promoting breast cancer growth
- PMID: 37880212
- PMCID: PMC10600207
- DOI: 10.1038/s41467-023-42458-1
Mutant p53 sustains serine-glycine synthesis and essential amino acids intake promoting breast cancer growth
Erratum in
-
Author Correction: Mutant p53 sustains serine-glycine synthesis and essential amino acids intake promoting breast cancer growth.Nat Commun. 2023 Nov 6;14(1):7129. doi: 10.1038/s41467-023-43018-3. Nat Commun. 2023. PMID: 37932286 Free PMC article. No abstract available.
Abstract
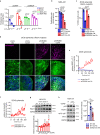
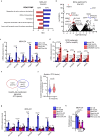
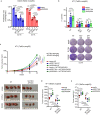
Reprogramming of amino acid metabolism, sustained by oncogenic signaling, is crucial for cancer cell survival under nutrient limitation. Here we discovered that missense mutant p53 oncoproteins stimulate de novo serine/glycine synthesis and essential amino acids intake, promoting breast cancer growth. Mechanistically, mutant p53, unlike the wild-type counterpart, induces the expression of serine-synthesis-pathway enzymes and L-type amino acid transporter 1 (LAT1)/CD98 heavy chain heterodimer. This effect is exacerbated by amino acid shortage, representing a mutant p53-dependent metabolic adaptive response. When cells suffer amino acids scarcity, mutant p53 protein is stabilized and induces metabolic alterations and an amino acid transcriptional program that sustain cancer cell proliferation. In patient-derived tumor organoids, pharmacological targeting of either serine-synthesis-pathway and LAT1-mediated transport synergizes with amino acid shortage in blunting mutant p53-dependent growth. These findings reveal vulnerabilities potentially exploitable for tackling breast tumors bearing missense TP53 mutations.
© 2023. Springer Nature Limited.
Conflict of interest statement
The authors declare no competing interests.
Figures




Similar articles
-
CircMYH9 drives colorectal cancer growth by regulating serine metabolism and redox homeostasis in a p53-dependent manner.Mol Cancer. 2021 Sep 8;20(1):114. doi: 10.1186/s12943-021-01412-9. Mol Cancer. 2021. PMID: 34496888 Free PMC article.
-
Platelet-derived growth factor stimulates LAT1 gene expression in vascular smooth muscle: role in cell growth.FASEB J. 2004 Apr;18(6):768-70. doi: 10.1096/fj.03-0886fje. Epub 2004 Feb 20. FASEB J. 2004. PMID: 14977877
-
Mutant p53 accumulation in human breast cancer is not an intrinsic property or dependent on structural or functional disruption but is regulated by exogenous stress and receptor status.J Pathol. 2014 Jul;233(3):238-46. doi: 10.1002/path.4356. Epub 2014 May 21. J Pathol. 2014. PMID: 24687952
-
Targeting mutant p53: a key player in breast cancer pathogenesis and beyond.Cell Commun Signal. 2024 Oct 10;22(1):484. doi: 10.1186/s12964-024-01863-9. Cell Commun Signal. 2024. PMID: 39390510 Free PMC article. Review.
-
The Role of Large Neutral Amino Acid Transporter (LAT1) in Cancer.Curr Cancer Drug Targets. 2019;19(11):863-876. doi: 10.2174/1568009619666190802135714. Curr Cancer Drug Targets. 2019. PMID: 31376820 Review.
Cited by
-
Targeting metabolism of breast cancer and its implications in T cell immunotherapy.Front Immunol. 2024 Apr 12;15:1381970. doi: 10.3389/fimmu.2024.1381970. eCollection 2024. Front Immunol. 2024. PMID: 38680483 Free PMC article. Review.
-
Understanding serine and glycine metabolism in cancer: a path towards precision medicine to improve patient's outcomes.Discov Oncol. 2024 Nov 13;15(1):652. doi: 10.1007/s12672-024-01544-6. Discov Oncol. 2024. PMID: 39538085 Free PMC article. Review.
-
Serine auxotrophy is a targetable vulnerability driven by PSAT1 suppression in AML.bioRxiv [Preprint]. 2025 May 14:2025.05.13.651470. doi: 10.1101/2025.05.13.651470. bioRxiv. 2025. PMID: 40462988 Free PMC article. Preprint.
-
Association between folate and glutamine metabolism and prognosis of kidney cancer.Front Nutr. 2025 Jan 31;11:1506967. doi: 10.3389/fnut.2024.1506967. eCollection 2024. Front Nutr. 2025. PMID: 39958609 Free PMC article.
-
Metabolic reprogramming in cancer and senescence.MedComm (2020). 2025 Mar 4;6(3):e70055. doi: 10.1002/mco2.70055. eCollection 2025 Mar. MedComm (2020). 2025. PMID: 40046406 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

