An ON-type direction-selective ganglion cell in primate retina
- PMID: 37880369
- PMCID: PMC10632142
- DOI: 10.1038/s41586-023-06659-4
An ON-type direction-selective ganglion cell in primate retina
Abstract
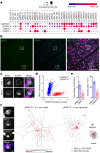
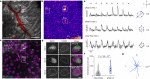
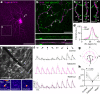
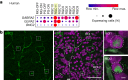
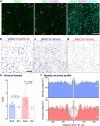
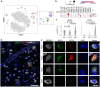
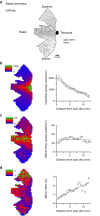
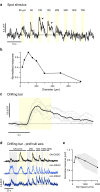
To maintain a stable and clear image of the world, our eyes reflexively follow the direction in which a visual scene is moving. Such gaze-stabilization mechanisms reduce image blur as we move in the environment. In non-primate mammals, this behaviour is initiated by retinal output neurons called ON-type direction-selective ganglion cells (ON-DSGCs), which detect the direction of image motion and transmit signals to brainstem nuclei that drive compensatory eye movements1. However, ON-DSGCs have not yet been identified in the retina of primates, raising the possibility that this reflex is mediated by cortical visual areas. Here we mined single-cell RNA transcriptomic data from primate retina to identify a candidate ON-DSGC. We then combined two-photon calcium imaging, molecular identification and morphological analysis to reveal a population of ON-DSGCs in the macaque retina. The morphology, molecular signature and GABA (γ-aminobutyric acid)-dependent mechanisms that underlie direction selectivity in primate ON-DSGCs are highly conserved with those in other mammals. We further identify a candidate ON-DSGC in human retina. The presence of ON-DSGCs in primates highlights the need to examine the contribution of subcortical retinal mechanisms to normal and aberrant gaze stabilization in the developing and mature visual system.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures

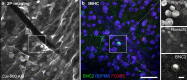
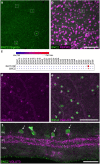
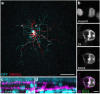
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

