Mass spectrometry quantifies target engagement for a KRASG12C inhibitor in FFPE tumor tissue
- PMID: 37880622
- PMCID: PMC10599008
- DOI: 10.1186/s12014-023-09435-8
Mass spectrometry quantifies target engagement for a KRASG12C inhibitor in FFPE tumor tissue
Abstract
Background: Quantification of drug-target binding is critical for confirming that drugs reach their intended protein targets, understanding the mechanism of action, and interpreting dose-response relationships. For covalent inhibitors, target engagement can be inferred by free target levels before and after treatment. Targeted mass spectrometry assays offer precise protein quantification in complex biological samples and have been routinely applied in pre-clinical studies to quantify target engagement in frozen tumor tissues for oncology drug development. However, frozen tissues are often not available from clinical trials so it is critical that assays are applicable to formalin-fixed, paraffin-embedded (FFPE) tissues in order to extend mass spectrometry-based target engagement studies into clinical settings.
Methods: Wild-type RAS and RASG12C was quantified in FFPE tissues by a highly optimized targeted mass spectrometry assay that couples high-field asymmetric waveform ion mobility spectrometry (FAIMS) and parallel reaction monitoring (PRM) with internal standards. In a subset of samples, technical reproducibility was evaluated by analyzing consecutive tissue sections from the same tumor block and biological variation was accessed among adjacent tumor regions in the same tissue section.
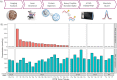
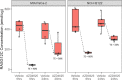
Results: Wild-type RAS protein was measured in 32 clinical non-small cell lung cancer tumors (622-2525 amol/µg) as measured by FAIMS-PRM mass spectrometry. Tumors with a known KRASG12C mutation (n = 17) expressed a wide range of RASG12C mutant protein (127-2012 amol/µg). The variation in wild-type RAS and RASG12C measurements ranged 0-18% CV across consecutive tissue sections and 5-20% CV among adjacent tissue regions. Quantitative target engagement was then demonstrated in FFPE tissues from 2 xenograft models (MIA PaCa-2 and NCI-H2122) treated with a RASG12C inhibitor (AZD4625).
Conclusions: This work illustrates the potential to expand mass spectrometry-based proteomics in preclinical and clinical oncology drug development through analysis of FFPE tumor biopsies.
Keywords: FFPE; Mass spectrometry; Target engagement; Targeted proteomics; Translational medicine.
© 2023. BioMed Central Ltd., part of Springer Nature.
Conflict of interest statement
All authors are employees of AstraZeneca and have stock ownership and/or stock options or interests in the company.
Figures

References
LinkOut - more resources
Full Text Sources
