A quantitative autonomous bioluminescence reporter system with a wide dynamic range for Plant Synthetic Biology
- PMID: 37882352
- PMCID: PMC10754000
- DOI: 10.1111/pbi.14146
A quantitative autonomous bioluminescence reporter system with a wide dynamic range for Plant Synthetic Biology
Abstract
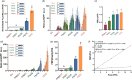
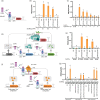
Plant Synthetic Biology aims to enhance the capacities of plants by designing and integrating synthetic gene circuits (SGCs). Quantitative reporting solutions that can produce quick, rich datasets affordably are necessary for SGC optimization. In this paper, we present a new, low-cost, and high-throughput reporter system for the quantitative measurement of gene expression in plants based on autonomous bioluminescence. This method eliminates the need for an exogenous supply of luciferase substrate by exploiting the entire Neonothopanus nambi fungal bioluminescence cyclic pathway to build a self-sustained reporter. The HispS gene, the pathway's limiting step, was set up as the reporter's transcriptional entry point as part of the new system's design, which significantly improved the output's dynamic range and brought it on par with that of the gold standard FLuc/RLuc reporter. Additionally, transient ratiometric measurements in N. benthamiana were made possible by the addition of an enhanced GFP as a normalizer. The performance of new NeoLuc/eGFP system was extensively validated with SGCs previously described, including phytohormone and optogenetic sensors. Furthermore, we employed NeoLuc/eGFP in the optimization of challenging SGCs, including new configurations for an agrochemical (copper) switch, a new blue optogenetic sensor, and a dual copper/red-light switch for tight regulation of metabolic pathways.
Keywords: Nicotiana benthamiana; Autonomous bioluminescence; ratiometric reporter system; transcriptional regulation.
© 2023 The Authors. Plant Biotechnology Journal published by Society for Experimental Biology and The Association of Applied Biologists and John Wiley & Sons Ltd.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




References
-
- Brophy, J.A.N. , Magallon, K.J. , Duan, L. , Zhong, V. , Ramachandran, P. , Kniazev, K. and Dinneny, J.R. (2022) Synthetic genetic circuits as a means of reprogramming plant roots. Science, 377, 747–751. - PubMed
-
- COVID vaccines grow on leaves (2021) COVID vaccines grow on leaves. Nat. Biotechnol. 39, 649. - PubMed
-
- Garcia‐Perez, E. , Diego‐Martin, B. , Quijano‐Rubio, A. , Moreno‐Giménez, E. , Selma, S. , Orzaez, D. and Vazquez‐Vilar, M. (2022) A copper switch for inducing CRISPR/Cas9‐based transcriptional activation tightly regulates gene expression in Nicotiana benthamiana . BMC Biotechnol. 22, 12. - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

