Comparison of phage-derived recombinases for genetic manipulation of Pseudomonas species
- PMID: 37882574
- PMCID: PMC10714826
- DOI: 10.1128/spectrum.03176-23
Comparison of phage-derived recombinases for genetic manipulation of Pseudomonas species
Abstract
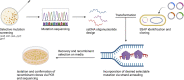
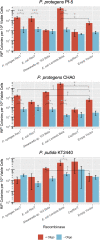
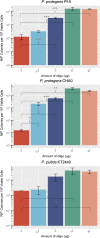
The Pseudomonas genus contains many members currently being investigated for applications in biodegradation, biopesticides, biocontrol, and synthetic biology. Though several strains have been identified with beneficial properties, chromosomal manipulations to further improve these strains for commercial applications have been limited due to the lack of efficient genetic tools that have been tested across this genus. Here, we test the recombineering efficiencies of five phage-derived recombinases across three biotechnologically relevant Pseudomonas strains: P. putida KT2440, P. protegens Pf-5, and P. protegens CHA0. These results demonstrate a method to generate targeted mutations quickly and efficiently across these strains, ideally introducing a method that can be implemented across the Pseudomonas genus and a strategy that may be applied to develop analogous systems in other nonmodel bacteria.
Keywords: Pseudomonas; SSAPs; homologous recombination; plant growth promoting rhizobacteria; recombineering.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

