Predicting Colorectal Cancer Survival Using Time-to-Event Machine Learning: Retrospective Cohort Study
- PMID: 37883174
- PMCID: PMC10636616
- DOI: 10.2196/44417
Predicting Colorectal Cancer Survival Using Time-to-Event Machine Learning: Retrospective Cohort Study
Abstract
Background: Machine learning (ML) methods have shown great potential in predicting colorectal cancer (CRC) survival. However, the ML models introduced thus far have mainly focused on binary outcomes and have not considered the time-to-event nature of this type of modeling.
Objective: This study aims to evaluate the performance of ML approaches for modeling time-to-event survival data and develop transparent models for predicting CRC-specific survival.
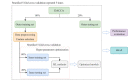
Methods: The data set used in this retrospective cohort study contains information on patients who were newly diagnosed with CRC between December 28, 2012, and December 27, 2019, at West China Hospital, Sichuan University. We assessed the performance of 6 representative ML models, including random survival forest (RSF), gradient boosting machine (GBM), DeepSurv, DeepHit, neural net-extended time-dependent Cox (or Cox-Time), and neural multitask logistic regression (N-MTLR) in predicting CRC-specific survival. Multiple imputation by chained equations method was applied to handle missing values in variables. Multivariable analysis and clinical experience were used to select significant features associated with CRC survival. Model performance was evaluated in stratified 5-fold cross-validation repeated 5 times by using the time-dependent concordance index, integrated Brier score, calibration curves, and decision curves. The SHapley Additive exPlanations method was applied to calculate feature importance.
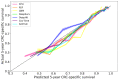
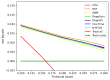
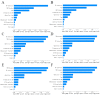
Results: A total of 2157 patients with CRC were included in this study. Among the 6 time-to-event ML models, the DeepHit model exhibited the best discriminative ability (time-dependent concordance index 0.789, 95% CI 0.779-0.799) and the RSF model produced better-calibrated survival estimates (integrated Brier score 0.096, 95% CI 0.094-0.099), but these are not statistically significant. Additionally, the RSF, GBM, DeepSurv, Cox-Time, and N-MTLR models have comparable predictive accuracy to the Cox Proportional Hazards model in terms of discrimination and calibration. The calibration curves showed that all the ML models exhibited good 5-year survival calibration. The decision curves for CRC-specific survival at 5 years showed that all the ML models, especially RSF, had higher net benefits than default strategies of treating all or no patients at a range of clinically reasonable risk thresholds. The SHapley Additive exPlanations method revealed that R0 resection, tumor-node-metastasis staging, and the number of positive lymph nodes were important factors for 5-year CRC-specific survival.
Conclusions: This study showed the potential of applying time-to-event ML predictive algorithms to help predict CRC-specific survival. The RSF, GBM, Cox-Time, and N-MTLR algorithms could provide nonparametric alternatives to the Cox Proportional Hazards model in estimating the survival probability of patients with CRC. The transparent time-to-event ML models help clinicians to more accurately predict the survival rate for these patients and improve patient outcomes by enabling personalized treatment plans that are informed by explainable ML models.
Keywords: SHAP; SHapley Additive exPlanations; colorectal cancer; machine learning; survival prediction; time-to-event.
©Xulin Yang, Hang Qiu, Liya Wang, Xiaodong Wang. Originally published in the Journal of Medical Internet Research (https://www.jmir.org), 26.10.2023.
Conflict of interest statement
Conflicts of Interest: None declared.
Figures
References
-
- Xi Y, Xu P. Global colorectal cancer burden in 2020 and projections to 2040. Transl Oncol. 2021 Oct;14(10):101174. doi: 10.1016/j.tranon.2021.101174. https://linkinghub.elsevier.com/retrieve/pii/S1936-5233(21)00166-2 S1936-5233(21)00166-2 - DOI - PMC - PubMed
-
- Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, Bray F. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021 Feb 04;:209–249. doi: 10.3322/caac.21660. doi: 10.3322/caac.21660. - DOI - PubMed
-
- Wang Y, Wang D, Ye X, Wang Y, Yin Y, Jin Y. A tree ensemble-based two-stage model for advanced-stage colorectal cancer survival prediction. Inf Sci. 2019 Feb;474:106–124. doi: 10.1016/j.ins.2018.09.046. - DOI
-
- Pourhoseingholi MA, Kheirian S, Zali MR. Comparison of basic and ensemble data mining methods in predicting 5-year survival of colorectal cancer patients. Acta Inform Med. 2017 Dec;25(4):254–258. doi: 10.5455/aim.2017.25.254-258. https://europepmc.org/abstract/MED/29284916 AIM-25-254 - DOI - PMC - PubMed
-
- Cox D. Regression models and life-tables. J R Stat Soc Series B Stat Methodol. 2018 Dec 05;34(2):187–202. doi: 10.1111/j.2517-6161.1972.tb00899.x. - DOI
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical

