Mammalian cells internalize bacteriophages and use them as a resource to enhance cellular growth and survival
- PMID: 37883333
- PMCID: PMC10602308
- DOI: 10.1371/journal.pbio.3002341
Mammalian cells internalize bacteriophages and use them as a resource to enhance cellular growth and survival
Abstract
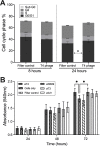
There is a growing appreciation that the direct interaction between bacteriophages and the mammalian host can facilitate diverse and unexplored symbioses. Yet the impact these bacteriophages may have on mammalian cellular and immunological processes is poorly understood. Here, we applied highly purified phage T4, free from bacterial by-products and endotoxins to mammalian cells and analyzed the cellular responses using luciferase reporter and antibody microarray assays. Phage preparations were applied in vitro to either A549 lung epithelial cells, MDCK-I kidney cells, or primary mouse bone marrow derived macrophages with the phage-free supernatant serving as a comparative control. Highly purified T4 phages were rapidly internalized by mammalian cells and accumulated within macropinosomes but did not activate the inflammatory DNA response TLR9 or cGAS-STING pathways. Following 8 hours of incubation with T4 phage, whole cell lysates were analyzed via antibody microarray that detected expression and phosphorylation levels of human signaling proteins. T4 phage application led to the activation of AKT-dependent pathways, resulting in an increase in cell metabolism, survival, and actin reorganization, the last being critical for macropinocytosis and potentially regulating a positive feedback loop to drive further phage internalization. T4 phages additionally down-regulated CDK1 and its downstream effectors, leading to an inhibition of cell cycle progression and an increase in cellular growth through a prolonged G1 phase. These interactions demonstrate that highly purified T4 phages do not activate DNA-mediated inflammatory pathways but do trigger protein phosphorylation cascades that promote cellular growth and survival. We conclude that mammalian cells are internalizing bacteriophages as a resource to promote cellular growth and metabolism.
Copyright: © 2023 Bichet et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
JJB has a patent application related to this work (WO2018129536A1).
Figures



References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

