Identification of conserved skeletal enhancers associated with craniosynostosis risk genes
- PMID: 37883470
- PMCID: PMC11070136
- DOI: 10.1093/hmg/ddad182
Identification of conserved skeletal enhancers associated with craniosynostosis risk genes
Abstract
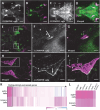
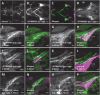
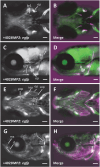
Craniosynostosis, defined by premature fusion of one or multiple cranial sutures, is a common congenital defect affecting more than 1/2000 infants and results in restricted brain expansion. Single gene mutations account for 15%-20% of cases, largely as part of a syndrome, but the majority are nonsyndromic with complex underlying genetics. We hypothesized that the two noncoding genomic regions identified by a GWAS for craniosynostosis contain distal regulatory elements for the risk genes BMPER and BMP2. To identify such regulatory elements, we surveyed conserved noncoding sequences from both risk loci for enhancer activity in transgenic Danio rerio. We identified enhancers from both regions that direct expression to skeletal tissues, consistent with the endogenous expression of bmper and bmp2. For each locus, we also found a skeletal enhancer that also contains a sequence variant associated with craniosynostosis risk. We examined the activity of each enhancer during craniofacial development and found that the BMPER-associated enhancer is active in the restricted region of cartilage closely associated with frontal bone initiation. The same enhancer is active in mouse skeletal tissues, demonstrating evolutionarily conserved activity. Using enhanced yeast one-hybrid assays, we identified transcription factors that bind each enhancer and observed differential binding between alleles, implicating multiple signaling pathways. Our findings help unveil the genetic mechanism of the two craniosynostosis risk loci. More broadly, our combined in vivo approach is applicable to many complex genetic diseases to build a link between association studies and specific genetic mechanisms.
Keywords: BMP signaling; conserved regulatory elements; craniosynostosis; enhanced yeast one-hybrid assay; zebrafish transgenesis.
© The Author(s) 2023. Published by Oxford University Press. All rights reserved. For Permissions, please email: journals.permissions@oup.com.
Figures


References
-
- Bandyopadhyay A, Yadav PS, Prashar P. BMP signaling in development and diseases: a pharmacological perspective. Biochem Pharmacol 2013;85:857–64. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

