Conformationally Restricted Glycopeptide Backbone Inhibits Gas-Phase H/D Scrambling between Glycan and Peptide Moieties
- PMID: 37883679
- PMCID: PMC10636759
- DOI: 10.1021/jacs.3c04068
Conformationally Restricted Glycopeptide Backbone Inhibits Gas-Phase H/D Scrambling between Glycan and Peptide Moieties
Abstract
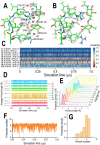
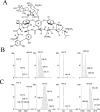
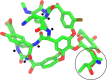
Protein glycosylation is a common post-translational modification on extracellular proteins. The conformational dynamics of several glycoproteins have been characterized by hydrogen/deuterium exchange mass spectrometry (HDX-MS). However, it is, in most cases, not possible to extract information about glycan conformation and dynamics due to the general difficulty of separating the deuterium content of the glycan from that of the peptide (in particular, for O-linked glycans). Here, we investigate whether the fragmentation of protonated glycopeptides by collision-induced dissociation (CID) can be used to determine the solution-specific deuterium content of the glycan. Central to this concept is that glycopeptides can undergo a facile loss of glycans upon CID, thereby allowing for the determination of their masses. However, an essential prerequisite is that hydrogen and deuterium (H/D) scrambling can be kept in check. Therefore, we have measured the degree of scrambling upon glycosidic bond cleavage in glycopeptides that differ in the conformational flexibility of their backbone and glycosylation pattern. Our results show that complete scrambling precedes the glycosidic bond cleavage in normal glycopeptides derived from a glycoprotein; i.e., all labile hydrogens have undergone positional randomization prior to loss of the glycan. In contrast, the glycosidic bond cleavage occurs without any scrambling in the glycopeptide antibiotic vancomycin, reflecting that the glycan cannot interact with the peptide moiety due to a conformationally restricted backbone as revealed by molecular dynamics simulations. Scrambling is also inhibited, albeit to a lesser degree, in the conformationally restricted glycopeptides ristocetin and its pseudoaglycone, demonstrating that scrambling depends on an intricate interplay between the flexibility and proximity of the glycan and the peptide backbone.
Conflict of interest statement
The authors declare no competing financial interest.
Figures



References
-
- Leth-Espensen K. Z.; Kristensen K. K.; Kumari A.; Winther A.-M. L.; Young S. G.; Jørgensen T. J. D.; Ploug M. The intrinsic instability of the hydrolase domain of lipoprotein lipase facilitates its inactivation by ANGPTL4-catalyzed unfolding. Proc. Natl. Acad. Sci. U.S.A. 2021, 118 (12), e2026650118 10.1073/pnas.2026650118. - DOI - PMC - PubMed
-
- Trelle M. B.; Pedersen S.; Østerlund E. C.; Madsen J. B.; Kristensen S. R.; Jørgensen T. J. D. An Asymmetric Runaway Domain Swap Antithrombin Dimer as a Key Intermediate for Polymerization Revealed by Hydrogen/Deuterium-Exchange Mass Spectrometry. Anal. Chem. 2017, 89 (1), 616–624. 10.1021/acs.analchem.6b02518. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

