A genetically encoded tool to increase cellular NADH/NAD+ ratio in living cells
- PMID: 37884806
- PMCID: PMC11045668
- DOI: 10.1038/s41589-023-01460-w
A genetically encoded tool to increase cellular NADH/NAD+ ratio in living cells
Abstract
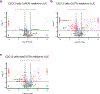
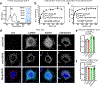
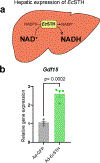
Impaired redox metabolism is a key contributor to the etiology of many diseases, including primary mitochondrial disorders, cancer, neurodegeneration and aging. However, mechanistic studies of redox imbalance remain challenging due to limited strategies that can perturb redox metabolism in various cellular or organismal backgrounds. Most studies involving impaired redox metabolism have focused on oxidative stress; consequently, less is known about the settings where there is an overabundance of NADH reducing equivalents, termed reductive stress. Here we introduce a soluble transhydrogenase from Escherichia coli (EcSTH) as a novel genetically encoded tool to promote reductive stress in living cells. When expressed in mammalian cells, EcSTH, and a mitochondrially targeted version (mitoEcSTH), robustly elevated the NADH/NAD+ ratio in a compartment-specific manner. Using this tool, we determined that metabolic and transcriptomic signatures of the NADH reductive stress are cellular background specific. Collectively, our novel genetically encoded tool represents an orthogonal strategy to promote reductive stress.
© 2023. The Author(s), under exclusive licence to Springer Nature America, Inc.
Conflict of interest statement
COMPETING INTERESTS:
VC is listed as an inventor on a patent application on the therapeutic uses of
Figures
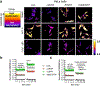

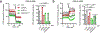

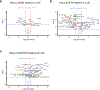
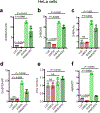







References
METHODS-ONLY REFERNCES:
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

