A citizen-science-enabled catalogue of the vaginal microbiome and associated factors
- PMID: 37884815
- PMCID: PMC10627828
- DOI: 10.1038/s41564-023-01500-0
A citizen-science-enabled catalogue of the vaginal microbiome and associated factors
Abstract
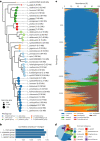
Understanding the composition and function of the vaginal microbiome is crucial for reproductive and overall health. Here we established the Isala citizen-science project to analyse the vaginal microbiomes of 3,345 women in Belgium (18-98 years) through self-sampling, 16S amplicon sequencing and extensive questionnaires. The overall vaginal microbiome composition was strongly tied to age, childbirth and menstrual cycle phase. Lactobacillus species dominated 78% of the vaginal samples. Specific bacterial taxa also showed to co-occur in modules based on network correlation analysis. Notably, the module containing Lactobacillus crispatus, Lactobacillus jensenii and Limosilactobacillus taxa was positively linked to oestrogen levels and contraceptive use and negatively linked to childbirth and breastfeeding. Other modules, named after abundant taxa (Gardnerella, Prevotella and Bacteroides), correlated with multiple partners, menopause, menstrual hygiene and contraceptive use. With this resource-rich vaginal microbiome map and associated health, life-course, lifestyle and dietary factors, we provide unique data and insights for follow-up clinical and mechanistic research.
© 2023. The Author(s).
Conflict of interest statement
S.L. declares to be a voluntary academic board member of the International Scientific Association on Probiotics and Prebiotics (ISAPP,
Figures




References
-
- Lash AF, Kaplan B. A study of Döderlein’ s vaginal bacillus. Oxford Univ. Press. 1928;38:333–340.
-
- Zheng J, et al. A taxonomic note on the genus Lactobacillus: description of 23 novel genera, emended description of the genus Lactobacillus beijerinck 1901, and union of Lactobacillaceae and Leuconostocaceae. Int. J. Syst. Evol. Microbiol. 2020;70:2782–2858. doi: 10.1099/ijsem.0.004107. - DOI - PubMed
MeSH terms
Substances
Grants and funding
- 852600/EC | EU Framework Programme for Research and Innovation H2020 | H2020 Priority Excellent Science | H2020 European Research Council (H2020 Excellent Science - European Research Council)
- DOCPRO 37054/Universiteit Antwerpen (University of Antwerp)
- 852600/EC | Horizon 2020 Framework Programme (EU Framework Programme for Research and Innovation H2020)
LinkOut - more resources
Full Text Sources

