Comparative analyses of the bacterial communities present in the spontaneously fermented milk products of Northeast India and West Africa
- PMID: 37886068
- PMCID: PMC10598763
- DOI: 10.3389/fmicb.2023.1166518
Comparative analyses of the bacterial communities present in the spontaneously fermented milk products of Northeast India and West Africa
Abstract
Introduction: Spontaneous fermentation of raw cow milk without backslopping is in practice worldwide as part of the traditional food culture, including "Doi" preparation in earthen pots in Northeast India, "Kindouri" of Niger and "Fanire" of Benin prepared in calabash vessels in West Africa. Very few reports are available about the differences in bacterial communities that evolved during the spontaneous mesophilic fermentation of cow milk in diverse geographical regions.
Methods: In this study, we used high throughput amplicon sequencing of bacterial 16S rRNA gene to investigate 44 samples of naturally fermented homemade milk products and compared the bacterial community structure of these foods, which are widely consumed in Northeast India and Western Africa.
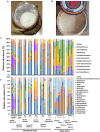
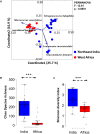
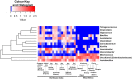
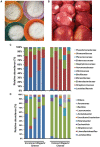
Results and discussion: The spontaneous milk fermentation shared the lactic acid bacteria, mainly belonging to Lactobacillaceae (Lactobacillus) and Streptococcaceae (Lactococcus) in these two geographically isolated regions. Indian samples showed a high bacterial diversity with the predominance of Acetobacteraceae (Gluconobacter and Acetobacter) and Leuconostoc, whereas Staphylococcaceae (Macrococcus) was abundant in the West African samples. However, the Wagashi cheese of Benin, prepared by curdling the milk with proteolytic leaf extract of Calotrophis procera followed by natural fermentation, contained Streptococcaceae (Streptococcus spp.) as the dominant bacteria. Our analysis also detected several potential pathogens, like Streptococcus infantarius an emerging infectious foodborne pathogen in Wagashi samples, an uncultured bacterium of Enterobacteriaceae in Kindouri and Fanire samples, and Clostridium spp. in the Doi samples of Northeast India. These findings will allow us to develop strategies to address the safety issues related to spontaneous milk fermentation and implement technological interventions for controlled milk fermentation by designing starter culture consortiums for the sustainable production of uniform quality products with desirable functional and organoleptic properties.
Keywords: Acetobacter; Gluconobacter; Lactobacillus; Lactococcus; Macrococcus caseolyticus; MiSeq amplicon sequencing; Streptococcus infantarius; spontaneously fermented milk products.
Copyright © 2023 Sessou, Keisam, Gagara, Komagbe, Farougou, Mahillon and Jeyaram.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
References
-
- Ampe F., ben Omar N., Moizan C., Wacher C., Guyot J. P. (1999). Polyphasic study of the spatial distribution of microorganisms in Mexican pozol, a fermented maise dough, demonstrates the need for cultivation-independent methods to investigate traditional fermentations. Appl. Environ. Microbiol. 65, 5464–5473. 10.1128/AEM.65.12.5464-5473.1999 - DOI - PMC - PubMed
-
- Anihouvi D. G. H., Henriet O., Kpoclou Y. E., Scippo M. L., Hounhouigan D. J., Anihouvi V. B., et al. (2021). Bacterial diversity of smoked and smoked-dried fish from West Africa: a metagenomic approach. J. Food Process. Preserv. 45, 1–47. 10.1111/jfpp.15919 - DOI
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

