Potential biomarkers in Japanese encephalitis from different hosts and geographical locations
- PMID: 37886150
- PMCID: PMC10599671
- DOI: 10.6026/97320630019611
Potential biomarkers in Japanese encephalitis from different hosts and geographical locations
Abstract
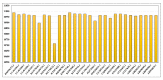
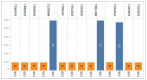
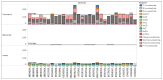
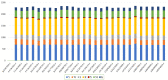
Japanese encephalitis (JE) is a single-stranded, mosquito-borne, positive-sense RNA flavivirus that causes one of the most severe encephalitides. There are treatments available for those who contact this illness; however, there are no known cures. This disease has a 30% fatality rate, and of the people who survive, 30-50% develops neurologic and psychiatric sequelae. The JE virus genome size is 10.98 kb and contains two coding DNA sequences (CDS), two genes, and 15 mature peptides; the CDS polyprotein is 10.3 kb. In this study, we used 29 genomics sequences of the JE virus reported from different countries and infecting different animals and analysed vast dimensions of the genomic annotation of JE comparatively to understand its evolutionary aspects. The extensive SNPs analysis revealed that KF907505.1, reported from Taiwan, has only three SNPs, similar to sequences reported from India. Repeat and polymorphism analyses revealed that the genome tends to be similar in most JE sequences.
Keywords: Japanese encephalitis virus; Single Nucleotide Polymorphism (SNPs); phobos; phylogenomics; transcription factors.
© 2023 Biomedical Informatics.
Conflict of interest statement
The authors declare there are no conflicts of interest.
Figures




References
LinkOut - more resources
Full Text Sources
