This is a preprint.
Multi-ancestry Polygenic Mechanisms of Type 2 Diabetes Elucidate Disease Processes and Clinical Heterogeneity
- PMID: 37886436
- PMCID: PMC10602111
- DOI: 10.21203/rs.3.rs-3399145/v1
Multi-ancestry Polygenic Mechanisms of Type 2 Diabetes Elucidate Disease Processes and Clinical Heterogeneity
Update in
-
Multi-ancestry polygenic mechanisms of type 2 diabetes.Nat Med. 2024 Apr;30(4):1065-1074. doi: 10.1038/s41591-024-02865-3. Epub 2024 Mar 5. Nat Med. 2024. PMID: 38443691 Free PMC article.
Abstract
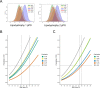
We identified genetic subtypes of type 2 diabetes (T2D) by analyzing genetic data from diverse groups, including non-European populations. We implemented soft clustering with 650 T2D-associated genetic variants, capturing known and novel T2D subtypes with distinct cardiometabolic trait associations. The twelve genetic clusters were distinctively enriched for single-cell regulatory regions. Polygenic scores derived from the clusters differed in distribution between ancestry groups, including a significantly higher proportion of lipodystrophy-related polygenic risk in East Asian ancestry. T2D risk was equivalent at a BMI of 30 kg/m2 in the European subpopulation and 24.2 (22.9-25.5) kg/m2 in the East Asian subpopulation; after adjusting for cluster-specific genetic risk, the equivalent BMI threshold increased to 28.5 (27.1-30.0) kg/m2 in the East Asian group, explaining about 75% of the difference in BMI thresholds. Thus, these multi-ancestry T2D genetic subtypes encompass a broader range of biological mechanisms and help explain ancestry-associated differences in T2D risk profiles.
Figures



References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources

