This is a preprint.
Population-based Risk of Psychiatric Disorders Associated with Recurrent CNVs
- PMID: 37886536
- PMCID: PMC10602037
- DOI: 10.1101/2023.09.04.23294975
Population-based Risk of Psychiatric Disorders Associated with Recurrent CNVs
Update in
-
Population-Based Risk of Psychiatric Disorders Associated With Recurrent Copy Number Variants.JAMA Psychiatry. 2024 Oct 1;81(10):957-966. doi: 10.1001/jamapsychiatry.2024.1453. JAMA Psychiatry. 2024. PMID: 38922630 Free PMC article.
Abstract
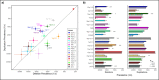
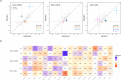
Recurrent copy number variants (rCNVs) are associated with increased risk of neuropsychiatric disorders but their pathogenic population-level impact is unknown. We provide population-based estimates of rCNV-associated risk of neuropsychiatric disorders for 34 rCNVs in the iPSYCH2015 case-cohort sample (n=120,247). Most observed significant increases in rCNV-associated risk for ADHD, autism or schizophrenia were moderate (HR:1.42-5.00), and risk estimates were highly correlated across these disorders, the most notable exception being high autism-associated risk with Prader-Willi/Angelman Syndrome duplications (HR=20.8). No rCNV was associated with significant increase in depression risk. Also, rCNV-associated risk was positively correlated with locus size and gene constraint. Comparison with published rCNV studies suggests that prevalence of some rCNVs is higher, and risk of psychiatric disorders lower, than previously estimated. In an era where genetics is increasingly being clinically applied, our results highlight the importance of population-based risk estimates for genetics-based predictions.
Figures


References
-
- Stefansson H., Meyer-Lindenberg A., Steinberg S., et al. CNVs conferring risk of autism or schizophrenia affect cognition in controls. Nature 505, 361–366 (2014). - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
