This is a preprint.
Prefrontal Cortical Regulation of REM Sleep
- PMID: 37886570
- PMCID: PMC10602053
- DOI: 10.21203/rs.3.rs-1417511/v1
Prefrontal Cortical Regulation of REM Sleep
Update in
-
Prefrontal cortical regulation of REM sleep.Nat Neurosci. 2023 Oct;26(10):1820-1832. doi: 10.1038/s41593-023-01398-1. Epub 2023 Sep 21. Nat Neurosci. 2023. PMID: 37735498
Abstract
Rapid-eye-movement (REM) sleep is accompanied by intense cortical activity, underlying its wake-like electroencephalogram (EEG). The neural activity inducing REM sleep is thought to originate from subcortical circuits in brainstem and hypothalamus. However, whether cortical neurons can also trigger REM sleep has remained unknown. Here, we show in mice that the medial prefrontal cortex (mPFC) strongly promotes REM sleep. Bidirectional optogenetic manipulations demonstrate that excitatory mPFC neurons promote REM sleep through their projections to the lateral hypothalamus (LH) and regulate phasic events, reflected in accelerated EEG theta oscillations and increased eye-movement density during REM sleep. Calcium imaging reveals that the majority of LH-projecting mPFC neurons are maximally activated during REM sleep and a subpopulation is recruited during phasic theta accelerations. Our results delineate a cortico-hypothalamic circuit for the top-down control of REM sleep and identify a critical role of the mPFC in regulating phasic events during REM sleep.
Conflict of interest statement
COMPETING INTERESTS The authors declare no competing interests.
Figures
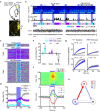



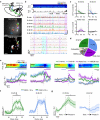

References
-
- Jouvet M. Recherches sur les structures nerveuses et les mécanismes responsables des différentes phases du sommeil physiologique. Arch Ital Biol 100, 125–206 (1962). - PubMed
-
- Maquet P. et al. Functional neuroanatomy of human rapid-eye-movement sleep and dreaming. Nature 383, 163–166 (1996). - PubMed
-
- Peever J. & Fuller P. M. The Biology of REM Sleep. Curr. Biol. 27, R1237–R1248 (2017). - PubMed
-
- Nofzinger E. A., Mintun M. A., Wiseman M., Kupfer D. J. & Moore R. Y. Forebrain activation in REM sleep: an FDG PET study. Brain Res. 770, 192–201 (1997). - PubMed
Publication types
LinkOut - more resources
Full Text Sources

