Identification of T-Cell Epitopes Using a Combined In-Silico and Experimental Approach in a Mouse Model for SARS-CoV-2
- PMID: 37886945
- PMCID: PMC10605721
- DOI: 10.3390/cimb45100502
Identification of T-Cell Epitopes Using a Combined In-Silico and Experimental Approach in a Mouse Model for SARS-CoV-2
Abstract
Following viral infection, T-cells are crucial for an effective immune response to intracellular pathogens, including respiratory viruses. During the COVID-19 pandemic, diverse assays were required in pre-clinical trials to evaluate the immune response following vaccination against SARS-CoV-2 and assess the response following exposure to the virus. To assess the nature and potency of the cellular response to infection or vaccination, a reliable and specific activity assay was needed. A cellular activity assay based on the presentation of short peptides (epitopes) allows the identification of T cell epitopes displayed on different alleles of the MHC, shedding light on the strength of the immune response towards antigens and aiding in antigen design for vaccination. In this report, we describe two approaches for scanning T cell epitopes on the surface glycoprotein of the SARS-CoV-2 (spike), which is utilized for attachment and entry and serves as an antigen in many vaccine candidates. We demonstrate that epitope scanning is feasible using peptide libraries or computational scanning combined with a cellular activity assay. Our scans identified four CD8 T cell epitopes, including one novel undescribed epitope. These epitopes enabled us to establish a reliable T-cell response assay, which was examined and used in various experimental mouse models for SARS-CoV-2 infection and vaccination. These approaches could potentially aid in future antigen design for vaccination and establish cellular activity assays against uncharacterized antigens of emerging pathogens.
Keywords: SARS-CoV-2; T-cell; T-cell epitope; immune response; mouse.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

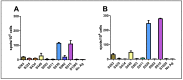

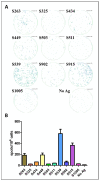
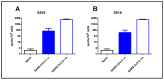
References
-
- Tarke A., Sidney J., Methot N., Yu E.D., Zhang Y., Dan J.M., Goodwin B., Rubiro P., Sutherland A., Wang E., et al. Impact of SARS-CoV-2 variants on the total CD4+ and CD8+ T cell reactivity in infected or vaccinated individuals. Cell Rep. Med. 2021;2:100355. doi: 10.1016/j.xcrm.2021.100355. - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

