Quantitative Imaging of Genetically Encoded Fluorescence Lifetime Biosensors
- PMID: 37887132
- PMCID: PMC10605767
- DOI: 10.3390/bios13100939
Quantitative Imaging of Genetically Encoded Fluorescence Lifetime Biosensors
Abstract
Genetically encoded fluorescence lifetime biosensors have emerged as powerful tools for quantitative imaging, enabling precise measurement of cellular metabolites, molecular interactions, and dynamic cellular processes. This review provides an overview of the principles, applications, and advancements in quantitative imaging with genetically encoded fluorescence lifetime biosensors using fluorescence lifetime imaging microscopy (go-FLIM). We highlighted the distinct advantages of fluorescence lifetime-based measurements, including independence from expression levels, excitation power, and focus drift, resulting in robust and reliable measurements compared to intensity-based approaches. Specifically, we focus on two types of go-FLIM, namely Förster resonance energy transfer (FRET)-FLIM and single-fluorescent protein (FP)-based FLIM biosensors, and discuss their unique characteristics and benefits. This review serves as a valuable resource for researchers interested in leveraging fluorescence lifetime imaging to study molecular interactions and cellular metabolism with high precision and accuracy.
Keywords: FLIM; FRET–FLIM; fluorescent proteins; genetically encoded fluorescence lifetime biosensors; quantitative imaging.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
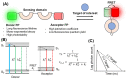
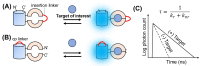
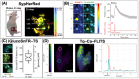

References
-
- Arai S., Kriszt R., Harada K., Looi L.-S., Matsuda S., Wongso D., Suo S., Ishiura S., Tseng Y.-H., Raghunath M., et al. RGB-Color Intensiometric Indicators to Visualize Spatiotemporal Dynamics of ATP in Single Cells. Angew. Chem. Int. Ed. 2018;57:10873–10878. doi: 10.1002/anie.201804304. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

