Identification of BRCC3 and BRCA1 as Regulators of TAZ Stability and Activity
- PMID: 37887275
- PMCID: PMC10605050
- DOI: 10.3390/cells12202431
Identification of BRCC3 and BRCA1 as Regulators of TAZ Stability and Activity
Abstract
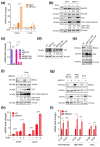
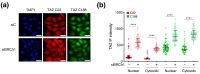
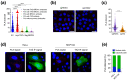
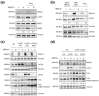
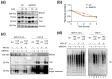
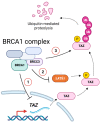
TAZ (WWTR1) is a transcriptional co-activator regulated by Hippo signaling, mechano-transduction, and G-protein couple receptors. Once activated, TAZ and its paralogue, YAP1, regulate gene expression programs promoting cell proliferation, survival, and differentiation, thus controlling embryonic development, tissue regeneration, and aging. YAP and TAZ are also frequently activated in tumors, particularly in poorly differentiated and highly aggressive malignancies. Yet, mutations of YAP/TAZ or of their upstream regulators do not fully account for their activation in cancer, raising the possibility that other upstream regulatory pathways, still to be defined, are altered in tumors. In this work, we set out to identify novel regulators of TAZ by means of a siRNA-based screen. We identified 200 genes able to modulate the transcriptional activity of TAZ, with prominence for genes implicated in cell-cell contact, cytoskeletal tension, cell migration, WNT signaling, chromatin remodeling, and interleukins and NF-kappaB signaling. Among these genes we identified was BRCC3, a component of the BRCA1 complex that guards genome integrity and exerts tumor suppressive activity during cancer development. The loss of BRCC3 or BRCA1 leads to an increased level and activity of TAZ. Follow-up studies indicated that the cytoplasmic BRCA1 complex controls the ubiquitination and stability of TAZ. This may suggest that, in tumors, inactivating mutations of BRCA1 may unleash cell transformation by activating the TAZ oncogene.
Keywords: BRCA1; BRCC3; TAZ; WWTR1; YAP1; post-translational regulation.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
Figures


References
-
- Liu C.Y., Zha Z.Y., Zhou X., Zhang H., Huang W., Zhao D., Li T., Chan S.W., Lim C.J., Hong W., et al. The hippo tumor pathway promotes TAZ degradation by phosphorylating a phosphodegron and recruiting the SCFbeta-TrCP E3 ligase. J. Biol. Chem. 2010;285:37159–37169. doi: 10.1074/jbc.M110.152942. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

