Identification and In Silico Characterization of a Conserved Peptide on Influenza Hemagglutinin Protein: A New Potential Antigen for Universal Influenza Vaccine Development
- PMID: 37887946
- PMCID: PMC10609762
- DOI: 10.3390/nano13202796
Identification and In Silico Characterization of a Conserved Peptide on Influenza Hemagglutinin Protein: A New Potential Antigen for Universal Influenza Vaccine Development
Abstract
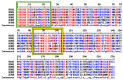
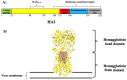
Antigenic changes in surface proteins of the influenza virus may cause the emergence of new variants that necessitate the reformulation of influenza vaccines every year. Universal influenza vaccine that relies on conserved regions can potentially be effective against all strains regardless of any antigenic changes and as a result, it can bring enormous public health impact and economic benefit worldwide. Here, a conserved peptide (HA288-107) on the stalk domain of hemagglutinin glycoprotein is identified among highly pathogenic influenza viruses. Five top-ranked B-cell and twelve T-cell epitopes were recognized by epitope mapping approaches and the corresponding Human Leukocyte Antigen alleles to T-cell epitopes showed high population coverage (>99%) worldwide. Moreover, molecular docking analysis indicated that VLMENERTL and WTYNAELLV epitopes have high binding affinity to the antigen-binding groove of the HLA-A*02:01 and HLA-A*68:02 molecules, respectively. Theoretical physicochemical properties of the peptide were assessed to ensure its thermostability and hydrophilicity. The results suggest that the HA288-107 peptide can be a promising antigen for universal influenza vaccine design. However, in vitro and in vivo analyses are needed to support and evaluate the effectiveness of the peptide as an immunogen for vaccine development.
Keywords: epitope mapping; hemagglutinin; immunoinformatic; nanoparticle; peptide-based vaccine; universal influenza vaccine.
Conflict of interest statement
The authors declare no conflict of interest.
Figures






Similar articles
-
In silico epitope-based vaccine design against influenza a neuraminidase protein: Computational analysis established on B- and T-cell epitope predictions.Saudi J Biol Sci. 2022 Jun;29(6):103283. doi: 10.1016/j.sjbs.2022.103283. Epub 2022 Apr 21. Saudi J Biol Sci. 2022. PMID: 35574284 Free PMC article.
-
Mutations in the Hemagglutinin Stalk Domain Do Not Permit Escape from a Protective, Stalk-Based Vaccine-Induced Immune Response in the Mouse Model.mBio. 2021 Feb 16;12(1):e03617-20. doi: 10.1128/mBio.03617-20. mBio. 2021. PMID: 33593972 Free PMC article.
-
A conserved multi-epitope-based vaccine designed by targeting hemagglutinin protein of highly pathogenic avian H5 influenza viruses.3 Biotech. 2020 Dec;10(12):546. doi: 10.1007/s13205-020-02544-3. Epub 2020 Nov 23. 3 Biotech. 2020. PMID: 33251084 Free PMC article.
-
Analysis of the conserved protective epitopes of hemagglutinin on influenza A viruses.Front Immunol. 2023 Feb 17;14:1086297. doi: 10.3389/fimmu.2023.1086297. eCollection 2023. Front Immunol. 2023. PMID: 36875062 Free PMC article. Review.
-
Progress on adenovirus-vectored universal influenza vaccines.Hum Vaccin Immunother. 2015;11(5):1209-22. doi: 10.1080/21645515.2015.1016674. Hum Vaccin Immunother. 2015. PMID: 25876176 Free PMC article. Review.
Cited by
-
Feasibility of Using a Type I IFN-Based Non-Animal Approach to Predict Vaccine Efficacy and Safety Profiles.Vaccines (Basel). 2024 May 27;12(6):583. doi: 10.3390/vaccines12060583. Vaccines (Basel). 2024. PMID: 38932312 Free PMC article. Review.
-
Innovative Nanomaterial Properties and Applications in Chemistry, Physics, Medicine, or Environment.Nanomaterials (Basel). 2024 Jan 9;14(2):145. doi: 10.3390/nano14020145. Nanomaterials (Basel). 2024. PMID: 38251110 Free PMC article.
References
-
- World Health Organization . World Health Organization Factsheet, Influenza (Seasonal) World Health Organization; Geneva, Switzerland: 2018.
-
- Centers for Disease Control and Prevention (CDC); National Center for Immunization and Respiratory Diseases (NCIRD) [(accessed on 1 June 2023)];2019 Available online: https://www.cdc.gov/flu/avianflu/influenza-a-virus-subtypes.htm.
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

