White-nose syndrome restructures bat skin microbiomes
- PMID: 37888992
- PMCID: PMC10714735
- DOI: 10.1128/spectrum.02715-23
White-nose syndrome restructures bat skin microbiomes
Abstract
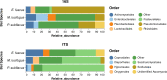
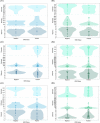
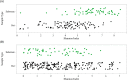
Inherent complexities in the composition of microbiomes can often preclude investigations of microbe-associated diseases. Instead of single organisms being associated with disease, community characteristics may be more relevant. Longitudinal microbiome studies of the same individual bats as pathogens arrive and infect a population are the ideal experiment but remain logistically challenging; therefore, investigations like our approach that are able to correlate invasive pathogens to alterations within a microbiome may be the next best alternative. The results of this study potentially suggest that microbiome-host interactions may determine the likelihood of infection. However, the contrasting relationship between Pd and the bacterial microbiomes of Myotis lucifugus and Perimyotis subflavus indicate that we are just beginning to understand how the bat microbiome interacts with a fungal invader such as Pd.
Keywords: Eptesicus fuscus; Myotis lucifugus; Perimyotis subflavus; Pseudogymnoascus destructans; bat populations; disease ecology; microbiome; white-nose syndrome.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Kong HH, Oh J, Deming C, Conlan S, Grice EA, Beatson MA, Nomicos E, Polley EC, Komarow HD, Program NCS, Murray PR, Turner ML, Segre JA. 2012. Temporal shifts in the skin microbiome associated with disease flares and treatment in children with atopic dermatitis. Genome Res 22:850–859. doi: 10.1101/gr.131029.111 - DOI - PMC - PubMed

