eRNAbase: a comprehensive database for decoding the regulatory eRNAs in human and mouse
- PMID: 37889077
- PMCID: PMC10767853
- DOI: 10.1093/nar/gkad925
eRNAbase: a comprehensive database for decoding the regulatory eRNAs in human and mouse
Abstract
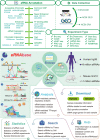
Enhancer RNAs (eRNAs) transcribed from distal active enhancers serve as key regulators in gene transcriptional regulation. The accumulation of eRNAs from multiple sequencing assays has led to an urgent need to comprehensively collect and process these data to illustrate the regulatory landscape of eRNAs. To address this need, we developed the eRNAbase (http://bio.liclab.net/eRNAbase/index.php) to store the massive available resources of human and mouse eRNAs and provide comprehensive annotation and analyses for eRNAs. The current version of eRNAbase cataloged 10 399 928 eRNAs from 1012 samples, including 858 human samples and 154 mouse samples. These eRNAs were first identified and uniformly processed from 14 eRNA-related experiment types manually collected from GEO/SRA and ENCODE. Importantly, the eRNAbase provides detailed and abundant (epi)genetic annotations in eRNA regions, such as super enhancers, enhancers, common single nucleotide polymorphisms, expression quantitative trait loci, transcription factor binding sites, CRISPR/Cas9 target sites, DNase I hypersensitivity sites, chromatin accessibility regions, methylation sites, chromatin interactions regions, topologically associating domains and RNA spatial interactions. Furthermore, the eRNAbase provides users with three novel analyses including eRNA-mediated pathway regulatory analysis, eRNA-based variation interpretation analysis and eRNA-mediated TF-target gene analysis. Hence, eRNAbase is a powerful platform to query, browse and visualize regulatory cues associated with eRNAs.
© The Author(s) 2023. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures



Similar articles
-
Pig-eRNAdb: a comprehensive enhancer and eRNA dataset of pigs.Sci Data. 2024 Feb 1;11(1):157. doi: 10.1038/s41597-024-02960-7. Sci Data. 2024. PMID: 38302497 Free PMC article.
-
eRNA-IDO: A One-stop Platform for Identification, Interactome Discovery, and Functional Annotation of Enhancer RNAs.Genomics Proteomics Bioinformatics. 2024 Oct 15;22(4):qzae059. doi: 10.1093/gpbjnl/qzae059. Genomics Proteomics Bioinformatics. 2024. PMID: 39178387 Free PMC article.
-
TRlnc: a comprehensive database for human transcriptional regulatory information of lncRNAs.Brief Bioinform. 2021 Mar 22;22(2):1929-1939. doi: 10.1093/bib/bbaa011. Brief Bioinform. 2021. PMID: 32047897
-
Enhancer RNA: What we know and what we can achieve.Cell Prolif. 2022 Apr;55(4):e13202. doi: 10.1111/cpr.13202. Epub 2022 Feb 16. Cell Prolif. 2022. PMID: 35170113 Free PMC article. Review.
-
Enhancer RNA: biogenesis, function, and regulation.Essays Biochem. 2020 Dec 7;64(6):883-894. doi: 10.1042/EBC20200014. Essays Biochem. 2020. PMID: 33034351 Review.
Cited by
-
Comprehensive Pan-Cancer Mutation Density Patterns in Enhancer RNA.Int J Mol Sci. 2023 Dec 30;25(1):534. doi: 10.3390/ijms25010534. Int J Mol Sci. 2023. PMID: 38203707 Free PMC article.
-
Intratumoral Heterogeneity and Immune Microenvironment in Hepatoblastoma Revealed by Single-Cell RNA Sequencing.J Cell Mol Med. 2025 Mar;29(6):e70482. doi: 10.1111/jcmm.70482. J Cell Mol Med. 2025. PMID: 40099956 Free PMC article.
-
LnCeCell 2.0: an updated resource for lncRNA-associated ceRNA networks and web tools based on single-cell and spatial transcriptomics sequencing data.Nucleic Acids Res. 2025 Jan 6;53(D1):D107-D115. doi: 10.1093/nar/gkae947. Nucleic Acids Res. 2025. PMID: 39470723 Free PMC article.
-
Deciphering the epigenetic role of long non-coding RNAs in mood disorders: Focus on human brain studies.Clin Transl Med. 2025 Mar;15(3):e70135. doi: 10.1002/ctm2.70135. Clin Transl Med. 2025. PMID: 40038891 Free PMC article. Review.
-
The miR-302 cluster-IRFs-IRF1AS axis regulates influenza A virus replication in a species-specific manner.mBio. 2025 Aug 13;16(8):e0137525. doi: 10.1128/mbio.01375-25. Epub 2025 Jul 8. mBio. 2025. PMID: 40626712 Free PMC article.
References
-
- Li W., Notani D., Rosenfeld M.G.. Enhancers as non-coding RNA transcription units: recent insights and future perspectives. Nat. Rev. Genet. 2016; 17:207–223. - PubMed
MeSH terms
Substances
Grants and funding
- 62171166/National Natural Science Foundation of China
- 20210002-1005 USCAT-2021-01/Affiliated Hospital of University of South China for Advanced Talents
- 2019M661311/China Postdoctoral Science Foundation
- 2023JJ30547/Natural Science Foundation of Hunan Province
- 20201920/Scientific Research Fund Project of Hunan Provincial Health Commission
LinkOut - more resources
Full Text Sources
Miscellaneous

