LSM14B is essential for oocyte meiotic maturation by regulating maternal mRNA storage and clearance
- PMID: 37889087
- PMCID: PMC10681746
- DOI: 10.1093/nar/gkad919
LSM14B is essential for oocyte meiotic maturation by regulating maternal mRNA storage and clearance
Abstract
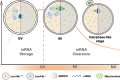
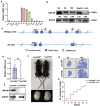
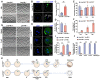
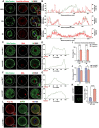
Fully grown oocytes remain transcriptionally quiescent, yet many maternal mRNAs are synthesized and retained in growing oocytes. We now know that maternal mRNAs are stored in a structure called the mitochondria-associated ribonucleoprotein domain (MARDO). However, the components and functions of MARDO remain elusive. Here, we found that LSM14B knockout prevents the proper storage and timely clearance of mRNAs (including Cyclin B1, Btg4 and other mRNAs that are translationally activated during meiotic maturation), specifically by disrupting MARDO assembly during oocyte growth and meiotic maturation. With decreased levels of storage and clearance, the LSM14B knockout oocytes failed to enter meiosis II, ultimately resulting in female infertility. Our results demonstrate the function of LSM14B in MARDO assembly, and couple the MARDO with mRNA clearance and oocyte meiotic maturation.
© The Author(s) 2023. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures

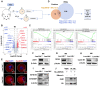
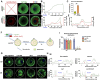
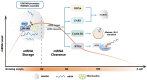
References
-
- Sha Q.Q., Zhang J., Fan H.Y.. A story of birth and death: mRNA translation and clearance at the onset of maternal-to-zygotic transition in mammals. Biol. Reprod. 2019; 101:579–590. - PubMed
-
- Liu Y., Lu X., Shi J., Yu X., Zhang X., Zhu K., Yi Z., Duan E., Li L.. BTG4 is a key regulator for maternal mRNA clearance during mouse early embryogenesis. J. Mol. Cell Biol. 2016; 8:366–368. - PubMed
-
- Jiang Y., Adhikari D., Li C., Zhou X.. Spatiotemporal regulation of maternal mRNAs during vertebrate oocyte meiotic maturation. Biol. Rev. Camb. Philos. Soc. 2023; 98:900–930. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
- 2022YFC2703200/National Key Research and Development Program of China
- 31988101/Basic Science Center Program of the NFSC
- 2021YFC2700200/National Key Research and Development Program of China
- 2019U001/Academic Promotion Programme of Shandong First Medical University
- 82101731/National Natural Science Foundation of China
- 2020RU001/Research Unit of Gametogenesis and Health of ART-Offspring, Chinese Academy of Medical Sciences
- 14103418/Research Grants Council of Hong Kong
- 2020ZLYS02/Shandong Provincial Key Research and Development Program
- 690598/CUHK-SDU Joint Laboratory on Reproductive Genetics of CUHK
- 2021ZDSYS16/Major Innovation Projects in Shandong Province
- ZR2021JQ27/Science Foundation for Distinguished Yong Scholars of Shandong
- tsqn202103192]/Taishan Scholars Program for Young Experts of Shandong Province

