Recognition and reconstruction of cell differentiation patterns with deep learning
- PMID: 37889897
- PMCID: PMC10631711
- DOI: 10.1371/journal.pcbi.1011582
Recognition and reconstruction of cell differentiation patterns with deep learning
Abstract
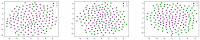
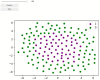
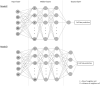
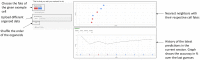
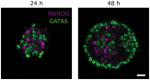
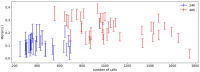
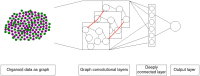
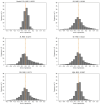
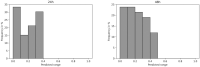
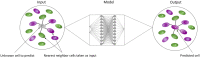
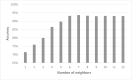
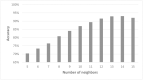
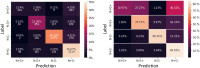
Cell lineage decisions occur in three-dimensional spatial patterns that are difficult to identify by eye. There is an ongoing effort to replicate such patterns using mathematical modeling. One approach uses long ranging cell-cell communication to replicate common spatial arrangements like checkerboard and engulfing patterns. In this model, the cell-cell communication has been implemented as a signal that disperses throughout the tissue. On the other hand, machine learning models have been developed for pattern recognition and pattern reconstruction tasks. We combined synthetic data generated by the mathematical model with spatial summary statistics and deep learning algorithms to recognize and reconstruct cell fate patterns in organoids of mouse embryonic stem cells. Application of Moran's index and pair correlation functions for in vitro and synthetic data from the model showed local clustering and radial segregation. To assess the patterns as a whole, a graph neural network was developed and trained on synthetic data from the model. Application to in vitro data predicted a low signal dispersion value. To test this result, we implemented a multilayer perceptron for the prediction of a given cell fate based on the fates of the neighboring cells. The results show a 70% accuracy of cell fate imputation based on the nine nearest neighbors of a cell. Overall, our approach combines deep learning with mathematical modeling to link cell fate patterns with potential underlying mechanisms.
Copyright: © 2023 Dirk et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures






References
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

