Deciphering sources of PET signals in the tumor microenvironment of glioblastoma at cellular resolution
- PMID: 37889970
- PMCID: PMC10610915
- DOI: 10.1126/sciadv.adi8986
Deciphering sources of PET signals in the tumor microenvironment of glioblastoma at cellular resolution
Abstract
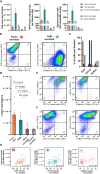
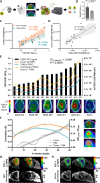
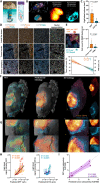
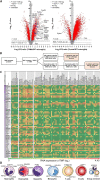
Various cellular sources hamper interpretation of positron emission tomography (PET) biomarkers in the tumor microenvironment (TME). We developed an approach of immunomagnetic cell sorting after in vivo radiotracer injection (scRadiotracing) with three-dimensional (3D) histology to dissect the cellular allocation of PET signals in the TME. In mice with implanted glioblastoma, translocator protein (TSPO) radiotracer uptake per tumor cell was higher compared to tumor-associated microglia/macrophages (TAMs), validated by protein levels. Translation of in vitro scRadiotracing to patients with glioma immediately after tumor resection confirmed higher single-cell TSPO tracer uptake of tumor cells compared to immune cells. Across species, cellular radiotracer uptake explained the heterogeneity of individual TSPO-PET signals. In consideration of cellular tracer uptake and cell type abundance, tumor cells were the main contributor to TSPO enrichment in glioblastoma; however, proteomics identified potential PET targets highly specific for TAMs. Combining cellular tracer uptake measures with 3D histology facilitates precise allocation of PET signals and serves to validate emerging novel TAM-specific radioligands.
Figures


References
-
- Pitt J. M., Marabelle A., Eggermont A., Soria J. C., Kroemer G., Zitvogel L., Targeting the tumor microenvironment: Removing obstruction to anticancer immune responses and immunotherapy. Ann. Oncol. 27, 1482–1492 (2016). - PubMed
-
- Kist de Ruijter L., van de Donk P. P., Hooiveld-Noeken J. S., Giesen D., Elias S. G., Lub-de Hooge M. N., Oosting S. F., Jalving M., Timens W., Brouwers A. H., Kwee T. C., Gietema J. A., Fehrmann R. S. N., Fine B. M., Sanabria Bohórquez S. M., Yadav M., Koeppen H., Jing J., Guelman S., Lin M. T., Mamounas M. J., Eastham J. R., Kimes P. K., Williams S. P., Ungewickell A., de Groot D. J. A., de Vries E. G. E., Whole-body CD8+ T cell visualization before and during cancer immunotherapy: A phase 1/2 trial. Nat. Med. 28, 2601–2610 (2022). - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

