Acute myeloid leukemias with UBTF tandem duplications are sensitive to menin inhibitors
- PMID: 37890156
- PMCID: PMC10873536
- DOI: 10.1182/blood.2023021359
Acute myeloid leukemias with UBTF tandem duplications are sensitive to menin inhibitors
Abstract
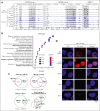
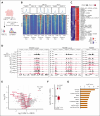
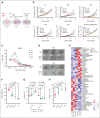
UBTF tandem duplications (UBTF-TDs) have recently emerged as a recurrent alteration in pediatric and adult acute myeloid leukemia (AML). UBTF-TD leukemias are characterized by a poor response to conventional chemotherapy and a transcriptional signature that mirrors NUP98-rearranged and NPM1-mutant AMLs, including HOX-gene dysregulation. However, the mechanism by which UBTF-TD drives leukemogenesis remains unknown. In this study, we investigated the genomic occupancy of UBTF-TD in transformed cord blood CD34+ cells and patient-derived xenograft models. We found that UBTF-TD protein maintained genomic occupancy at ribosomal DNA loci while also occupying genomic targets commonly dysregulated in UBTF-TD myeloid malignancies, such as the HOXA/HOXB gene clusters and MEIS1. These data suggest that UBTF-TD is a gain-of-function alteration that results in mislocalization to genomic loci dysregulated in UBTF-TD leukemias. UBTF-TD also co-occupies key genomic loci with KMT2A and menin, which are known to be key partners involved in HOX-dysregulated leukemias. Using a protein degradation system, we showed that stemness, proliferation, and transcriptional signatures are dependent on sustained UBTF-TD localization to chromatin. Finally, we demonstrate that primary cells from UBTF-TD leukemias are sensitive to the menin inhibitor SNDX-5613, resulting in markedly reduced in vitro and in vivo tumor growth, myeloid differentiation, and abrogation of the UBTF-TD leukemic expression signature. These findings provide a viable therapeutic strategy for patients with this high-risk AML subtype.
© 2024 American Society of Hematology. Published by Elsevier Inc. Licensed under Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International (CC BY-NC-ND 4.0), permitting only noncommercial, nonderivative use with attribution. All other rights reserved.
Conflict of interest statement
Conflict-of-interest disclosure: J.E.R. has been a consultant for Biomea Fusion. The remaining authors declare no competing financial interests.
Figures




Comment in
-
Menin dependence: UBTF-ITD AML joins the club.Blood. 2024 Feb 15;143(7):567-569. doi: 10.1182/blood.2023023041. Blood. 2024. PMID: 38358850 No abstract available.
References
-
- Sanij E, Hannan RD. The role of UBF in regulating the structure and dynamics of transcriptionally active rDNA chromatin. Epigenetics. 2009;4(6):374–382. - PubMed
-
- Moss T, Mars JC, Tremblay MG, Sabourin-Felix M. The chromatin landscape of the ribosomal RNA genes in mouse and human. Chromosome Res. 2019;27(1-2):31–40. - PubMed
-
- Kaburagi T, Shiba N, Yamato G, et al. UBTF-Internal tandem duplication as a novel poor prognostic factor in pediatric acute myeloid leukemia. Genes Chromosomes Cancer. 2023;62(4):202–209. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

