HSF1 Inhibits Antitumor Immune Activity in Breast Cancer by Suppressing CCL5 to Block CD8+ T-cell Recruitment
- PMID: 37890164
- PMCID: PMC10790131
- DOI: 10.1158/0008-5472.CAN-23-0902
HSF1 Inhibits Antitumor Immune Activity in Breast Cancer by Suppressing CCL5 to Block CD8+ T-cell Recruitment
Abstract
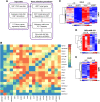
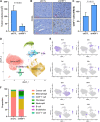
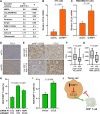
Heat shock factor 1 (HSF1) is a stress-responsive transcription factor that promotes cancer cell malignancy. To provide a better understanding of the biological processes regulated by HSF1, here we developed an HSF1 activity signature (HAS) and found that it was negatively associated with antitumor immune cells in breast tumors. Knockdown of HSF1 decreased breast tumor size and caused an influx of several antitumor immune cells, most notably CD8+ T cells. Depletion of CD8+ T cells rescued the reduction in growth of HSF1-deficient tumors, suggesting HSF1 prevents CD8+ T-cell influx to avoid immune-mediated tumor killing. HSF1 suppressed expression of CCL5, a chemokine for CD8+ T cells, and upregulation of CCL5 upon HSF1 loss significantly contributed to the recruitment of CD8+ T cells. These findings indicate that HSF1 suppresses antitumor immune activity by reducing CCL5 to limit CD8+ T-cell homing to breast tumors and prevent immune-mediated destruction, which has implications for the lack of success of immune modulatory therapies in breast cancer.
Significance: The stress-responsive transcription factor HSF1 reduces CD8+ T-cell infiltration in breast tumors to prevent immune-mediated killing, indicating that cellular stress responses affect tumor-immune interactions and that targeting HSF1 could improve immunotherapies.
©2023 The Authors; Published by the American Association for Cancer Research.
Figures





References
-
- American Cancer Society. Cancer Facts & Figures 2023. Atlanta, GA; 2023. Available from: https://www.cancer.org/content/dam/cancer-org/research/cancer-facts-and-....
-
- Gaynor N, Crown J, Collins DM. Immune checkpoint inhibitors: key trials and an emerging role in breast cancer. Semin Cancer Biol 2022;79:44–57. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

