Mowat-Wilson syndrome factor ZEB2 controls early formation of human neural crest through BMP signaling modulation
- PMID: 37890485
- PMCID: PMC10679662
- DOI: 10.1016/j.stemcr.2023.10.002
Mowat-Wilson syndrome factor ZEB2 controls early formation of human neural crest through BMP signaling modulation
Abstract
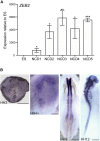
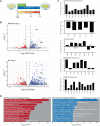
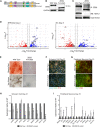
Mowat-Wilson syndrome is caused by mutations in ZEB2, with patients exhibiting characteristics indicative of neural crest (NC) defects. We examined the contribution of ZEB2 to human NC formation using a model based on human embryonic stem cells. We found ZEB2 to be one of the earliest factors expressed in prospective human NC, and knockdown revealed a role for ZEB2 in establishing the NC state while repressing pre-placodal and non-neural ectoderm genes. Examination of ZEB2 N-terminal mutant NC cells demonstrates its requirement for the repression of enhancers in the NC gene network and proper NC cell terminal differentiation into osteoblasts and peripheral neurons and neuroglia. This ZEB2 mutation causes early misexpression of BMP signaling ligands, which can be rescued by the attenuation of BMP. Our findings suggest that ZEB2 regulates early human NC specification by modulating proper BMP signaling and further elaborate the molecular defects underlying Mowat-Wilson syndrome.
Keywords: ATAC-seq; NuRD; RNA-seq; Schwann cells; ZEB2; epigenetics; human embryonic stem cells; neural crest; osteoblasts; peripheral neurons.
Copyright © 2023 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures


References
-
- Acloque H., Ocaña O.H., Abad D., Stern C.D., Nieto M.A. Snail2 and Zeb2 repress P-cadherin to define embryonic territories in the chick embryo. Development. 2017;144:649–656. - PubMed
-
- Adam M.P., Schelley S., Gallagher R., Brady A.N., Barr K., Blumberg B., Shieh J.T.C., Graham J., Slavotinek A., Martin M., et al. Clinical features and management issues in Mowat–Wilson syndrome. Am. J. Med. Genet. 2006;140:2730–2741. - PubMed
-
- Ahringer J. NuRD and SIN3 histone deacetylase complexes in development. Trends Genet. 2000;16:351–356. - PubMed
-
- Basch M.L., Bronner-Fraser M., García-Castro M.I. Specification of the neural crest occurs during gastrulation and requires Pax7. Nature. 2006;441:218–222. - PubMed
-
- Cacheux V., Dastot-Le Moal F., Kääriäinen H., Bondurand N., Rintala R., Boissier B., Wilson M., Mowat D., Goossens M. Loss-of-function mutations in SIP1 Smad interacting protein 1 result in a syndromic Hirschsprung disease. Hum. Mol. Genet. 2001;10:1503–1510. - PubMed
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

