Promoter-proximal introns impact recombinant amylase expression in Saccharomyces cerevisiae
- PMID: 37891015
- PMCID: PMC10647015
- DOI: 10.1093/femsyr/foad047
Promoter-proximal introns impact recombinant amylase expression in Saccharomyces cerevisiae
Abstract
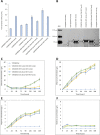
Consolidated bioprocessing (CBP) of starch requires recombinant Saccharomyces cerevisiae strains that produce raw starch-degrading enzymes and ferment the resultant sugars to ethanol in a single step. In this study, the native S. cerevisiae COX4 and RPS25A promoter-proximal introns were evaluated for enhanced expression of amylase genes (ateA, temA or temG_Opt) under the control of an S. cerevisiae promoter (ENO1P, TEF1P, TDH3P, or HXT7P). The results showed that different promoters and promoter-intron combinations differentially affected recombinant amylase production: ENO1P-COX4i and TDH3P-RPS25Ai were the best promoters for AteA, followed closely by HXT7P. The latter was also the best promoter for TemA and TemG production, followed closely by TDH3P-RPS25Ai for both these enzymes. Introducing promoter-proximal introns increased amylase activity up to 62% in Y294[ENO-COX-AteA] and Y294[TDH3-RPS-TemA], a significant improvement relative to the intron-less promoters. Strains co-expressing both an α-amylase and glucoamylase genes yielded up to 56 g/L ethanol from 20% w/v raw starch, with a higher carbon conversion observed with strains co-expressing TDH3P-RPS25Ai-temG_Opt than HXT7P-temG_Opt. The study showed that promoter-proximal introns can enhance amylase activity in S. cerevisiae and suggest that these alternative cassettes may also be considered for expression in more efficient ethanol-producing industrial yeast strains for raw starch CBP.
Keywords: intron-mediated enhancement; promoter-proximal introns; recombinant amylase; starch-based biofuels.
© The Author(s) 2023. Published by Oxford University Press on behalf of FEMS.
Conflict of interest statement
The authors declare that they have no conflict of interest related to the content of this paper.
Figures


References
-
- Ares Yeast Database (version 4.1) . 2011; Available at: http://intron.ucsc.edu/yeast4.1/. (20 July 2020, date last accessed).
-
- Blazeck J, Garg R, Reed Bet al. Controlling promoter strength and regulation in Saccharomyces cerevisiae using synthetic hybrid promoters. Biotech & Bioengineering. 2012;109:2884–95. - PubMed
-
- Cho KM, Yoo YJ, Kang HS. δ-Integration of endo/exo-glucanase and β-glucosidase genes into the yeast chromosome for direct conversion of cellulose to ethanol. Enzyme Microb Technol. 1999;25:23–30.
-
- Cripwell RA, Favaro L, Viljoen-Bloom Met al. Consolidated bioprocessing of raw starch to ethanol by Saccharomyces cerevisiae: achievements and challenges. Biotechnol Adv. 2020;42:107579. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

