SET-PP2A complex as a new therapeutic target in KMT2A (MLL) rearranged AML
- PMID: 37891368
- PMCID: PMC10709139
- DOI: 10.1038/s41388-023-02840-1
SET-PP2A complex as a new therapeutic target in KMT2A (MLL) rearranged AML
Abstract
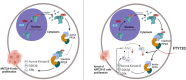
KMT2A-rearranged (KMT2A-R) is an aggressive and chemo-refractory acute leukemia which mostly affects children. Transcriptomics-based characterization and chemical interrogation identified kinases as key drivers of survival and drug resistance in KMT2A-R leukemia. In contrast, the contribution and regulation of phosphatases is unknown. In this study we uncover the essential role and underlying mechanisms of SET, the endogenous inhibitor of Ser/Thr phosphatase PP2A, in KMT2A-R-leukemia. Investigation of SET expression in acute myeloid leukemia (AML) samples demonstrated that SET is overexpressed, and elevated expression of SET is correlated with poor prognosis and with the expression of MEIS and HOXA genes in AML patients. Silencing SET specifically abolished the clonogenic ability of KMT2A-R leukemic cells and the transcription of KMT2A targets genes HOXA9 and HOXA10. Subsequent mechanistic investigations showed that SET interacts with both KMT2A wild type and fusion proteins, and it is recruited to the HOXA10 promoter. Pharmacological inhibition of SET by FTY720 disrupted SET-PP2A interaction leading to cell cycle arrest and increased sensitivity to chemotherapy in KMT2A-R-leukemic models. Phospho-proteomic analyses revealed that FTY720 reduced the activity of kinases regulated by PP2A, including ERK1, GSK3β, AURB and PLK1 and led to suppression of MYC, supporting the hypothesis of a feedback loop among PP2A, AURB, PLK1, MYC, and SET. Our findings illustrate that SET is a novel player in KMT2A-R leukemia and they provide evidence that SET antagonism could serve as a novel strategy to treat this aggressive leukemia.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures







References
-
- Dawson MA, Prinjha RK, Dittmann A, Nature GG. Inhibition of BET recruitment to chromatin as an effective treatment for MLL-fusion leukaemia. Inhibition of BET recruitment to chromatin as an effective treatment for MLL-fusion leukaemia. Nature. 2011;478:529–33. doi: 10.1038/nature10509. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

