Resurgence of SARS-CoV-2 Delta after Omicron variant superinfection in an immunocompromised pediatric patient
- PMID: 37891657
- PMCID: PMC10604949
- DOI: 10.1186/s12985-023-02186-w
Resurgence of SARS-CoV-2 Delta after Omicron variant superinfection in an immunocompromised pediatric patient
Abstract
Background: Persistent SARS-CoV-2 infection in immunocompromised hosts is thought to contribute to viral evolution by facilitating long-term natural selection and viral recombination in cases of viral co-infection or superinfection. However, there are limited data on the longitudinal intra-host population dynamics of SARS-CoV-2 co-infection/superinfection, especially in pediatric populations. Here, we report a case of Delta-Omicron superinfection in a hospitalized, immunocompromised pediatric patient.
Methods: We conducted Illumina whole genome sequencing (WGS) for longitudinal specimens to investigate intra-host dynamics of SARS-CoV-2 strains. Topoisomerase PCR cloning of Spike open-reading frame and Sanger sequencing of samples was performed for four specimens to validate the findings. Analysis of publicly available SARS-CoV-2 sequence data was performed to investigate the co-circulation and persistence of SARS-CoV-2 variants.
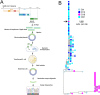
Results: Results of WGS indicate the patient was initially infected with the SARS-CoV-2 Delta variant before developing a SARS-CoV-2 Omicron variant superinfection, which became predominant. Shortly thereafter, viral loads decreased below the level of detection before resurgence of the original Delta variant with no residual trace of Omicron. After 54 days of persistent infection, the patient tested negative for SARS-CoV-2 but ultimately succumbed to a COVID-19-related death. Despite protracted treatment with remdesivir, no antiviral resistance mutations emerged. These results indicate a unique case of persistent SARS-CoV-2 infection with the Delta variant interposed by a transient superinfection with the Omicron variant. Analysis of publicly available sequence data suggests the persistence and ongoing evolution of Delta subvariants despite the global predominance of Omicron, potentially indicative of continued transmission in an unknown population or niche.
Conclusion: A better understanding of SARS-CoV-2 intra-host population dynamics, persistence, and evolution during co-infections and/or superinfections will be required to continue optimizing patient care and to better predict the emergence of new variants of concern.
Keywords: Delta; Omicron; SARS-CoV-2; Superinfection; Variants of concern.
© 2023. The Author(s).
Conflict of interest statement
J.F.H. has received research support, paid to Northwestern University, from Gilead Sciences and is a paid consultant for Merck. W.J.M. has received research support, paid to Lurie Children’s, from Ansun Biopharma, Astellas Pharma, AstraZeneca, Eli Lilly and Company, Enanta Pharmaceuticals, F. Hoffmann-La Roche, Gilead Sciences, Janssen Biotech, Karius, Inc., Melinta Therapeutics, Inc., Merck, Moderna, Nabriva Therapeutics, plc, Paratek Pharmaceuticals, Inc., Pfizer, and Tetraphase Pharmaceuticals, Inc. and has been a paid consultant for AstraZeneca, DiaSorin Molecular, Invivyd, and Sanofi Pasteur. All other authors declare no conflicts of interest.
Figures



References
-
- Tegally H, et al. Detection of a SARS-CoV-2 variant of concern in South Africa. Nature. Apr 2021;592(7854):438–43. 10.1038/s41586-021-03402-9. - PubMed
Publication types
MeSH terms
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

