Deep Neural Network-Based Automatic Dicentric Chromosome Detection Using a Model Pretrained on Common Objects
- PMID: 37892012
- PMCID: PMC10606160
- DOI: 10.3390/diagnostics13203191
Deep Neural Network-Based Automatic Dicentric Chromosome Detection Using a Model Pretrained on Common Objects
Abstract
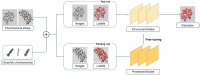
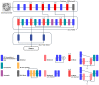
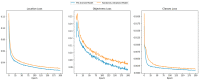
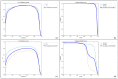
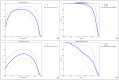
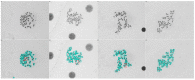
Dicentric chromosome assay (DCA) is one of the cytogenetic dosimetry methods where the absorbed dose is estimated by counting the number of dicentric chromosomes, which is a major radiation-induced change in DNA. However, DCA is a time-consuming task and requires technical expertise. In this study, a neural network was applied for automating the DCA. We used YOLOv5, a one-stage detection algorithm, to mitigate these limitations by automating the estimation of the number of dicentric chromosomes in chromosome metaphase images. YOLOv5 was pretrained on common object datasets. For training, 887 augmented chromosome images were used. We evaluated the model using validation and test datasets with 380 and 300 images, respectively. With pretrained parameters, the trained model detected chromosomes in the images with a maximum F1 score of 0.94 and a mean average precision (mAP) of 0.961. Conversely, when the model was randomly initialized, the training performance decreased, with a maximum F1 score and mAP of 0.82 and 0.873%, respectively. These results confirm that the model could effectively detect dicentric chromosomes in an image. Consequently, automatic DCA is expected to be conducted based on deep learning for object detection, requiring a relatively small amount of chromosome data for training using the pretrained network.
Keywords: chromosome metaphases image; cytogenetic dosimetry; deep learning; dicentric chromosome assay; object detection; transfer learning; you only look once.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Similar articles
-
Feasibility Study on Automatic Interpretation of Radiation Dose Using Deep Learning Technique for Dicentric Chromosome Assay.Radiat Res. 2021 Feb 1;195(2):163-172. doi: 10.1667/RADE-20-00167.1. Radiat Res. 2021. PMID: 33316052
-
Improving the accuracy of dose estimates from automatically scored dicentric chromosomes by accounting for chromosome number.Int J Radiat Biol. 2020 Dec;96(12):1571-1584. doi: 10.1080/09553002.2020.1829152. Epub 2020 Oct 16. Int J Radiat Biol. 2020. PMID: 33001765
-
Radiation dose estimation with multiple artificial neural networks in dicentric chromosome assay.Int J Radiat Biol. 2024;100(6):865-874. doi: 10.1080/09553002.2024.2338531. Epub 2024 Apr 30. Int J Radiat Biol. 2024. PMID: 38687685
-
High-precision automatic identification method for dicentric chromosome images using two-stage convolutional neural network.Sci Rep. 2023 Feb 6;13(1):2124. doi: 10.1038/s41598-023-28456-9. Sci Rep. 2023. PMID: 36746997 Free PMC article.
-
Development of electronic training and telescoring tools to increase the surge capacity of dicentric chromosome scorers for radiological/nuclear mass casualty incidents.Appl Radiat Isot. 2019 Feb;144:111-117. doi: 10.1016/j.apradiso.2018.12.005. Epub 2018 Dec 5. Appl Radiat Isot. 2019. PMID: 30572199 Free PMC article.
Cited by
-
Automated system for establishing standard radiation dose-response curves and dose estimation for the Korean population.Sci Rep. 2025 Mar 27;15(1):10639. doi: 10.1038/s41598-025-94678-8. Sci Rep. 2025. PMID: 40148494 Free PMC article.
-
Single-Cell Sequencing: An Emerging Tool for Biomarker Development in Nuclear Emergencies and Radiation Oncology.Cancers (Basel). 2025 May 28;17(11):1801. doi: 10.3390/cancers17111801. Cancers (Basel). 2025. PMID: 40507282 Free PMC article. Review.
References
Grants and funding
LinkOut - more resources
Full Text Sources

