Treatment-Emergent Cilgavimab Resistance Was Uncommon in Vaccinated Omicron BA.4/5 Outpatients
- PMID: 37892220
- PMCID: PMC10605390
- DOI: 10.3390/biom13101538
Treatment-Emergent Cilgavimab Resistance Was Uncommon in Vaccinated Omicron BA.4/5 Outpatients
Abstract
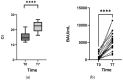
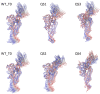
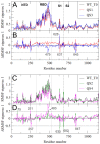
Mutations in the SARS-CoV-2 Spike glycoprotein can affect monoclonal antibody efficacy. Recent findings report the occurrence of resistant mutations in immunocompromised patients after tixagevimab/cilgavimab treatment. More recently, the Food and Drug Agency revoked the authorization for tixagevimab/cilgavimab, while this monoclonal antibody cocktail is currently recommended by the European Medical Agency. We retrospectively reviewed 22 immunocompetent patients at high risk for disease progression who received intramuscular tixagevimab/cilgavimab as early COVID-19 treatment and presented a prolonged high viral load. Complete SARS-CoV-2 genome sequences were obtained for a deep investigation of mutation frequencies in Spike protein before and during treatment. At seven days, only one patient showed evidence of treatment-emergent cilgavimab resistance. Quasispecies analysis revealed two different deletions on the Spike protein (S:del138-144 or S:del141-145) in combination with the resistance S:K444N mutation. The structural and dynamic impact of the two quasispecies was characterized by using molecular dynamics simulations, showing the conservation of the principal functional movements in the mutated systems and their capabilities to alter the structure and dynamics of the RBD, responsible for the interaction with the ACE2 human receptor. Our study underlines the importance of prompting an early virological investigation to prevent drug resistance or clinical failures in immunocompetent patients.
Keywords: SARS-CoV-2; Spike; cilgavimab; deletion; mAbs; mutations; quasispecies; tixagevimab.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
Figures

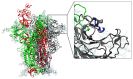
References
-
- FDA Announces Evusheld Is Not Currently Authorized for Emergency Use in the U.S. [(accessed on 3 February 2023)]; Available online: https://www.fda.gov/drugs/drug-safety-and-availability/fda-announces-evu....
-
- European Medicines Agency, Evusheld. [(accessed on 3 February 2023)]. Available online: https://www.ema.europa.eu/en/medicines/human/EPAR/evusheld.
-
- FDA Fact Sheet for Healthcare Providers: Emergency Use Authorization for Evusheld™ (Tixagevimab Co-Packaged with Cilgavimab) [(accessed on 3 February 2023)];2021 Available online: https://www.fda.gov/media/154701/download.
-
- Ordaya E.E., Vergidis P., Razonable R.R., Yao J.D., Beam E. Genotypic and predicted phenotypic analysis of SARS-CoV-2 Omicron subvariants in immunocompromised patients with COVID-19 following tixagevimab-cilgavimab prophylaxis. J. Clin. Virol. 2023;160:105382. doi: 10.1016/j.jcv.2023.105382. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

