Improvement of Locomotion Caused by Lactococcus lactis subsp. lactis in the Model Organism Caenorhabditis elegans
- PMID: 37892557
- PMCID: PMC10610199
- DOI: 10.3390/nu15204482
Improvement of Locomotion Caused by Lactococcus lactis subsp. lactis in the Model Organism Caenorhabditis elegans
Abstract
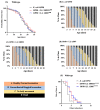
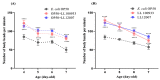
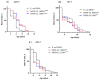
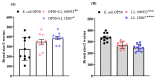
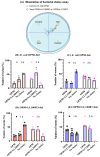
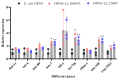
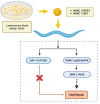
Lactococcus lactis subsp. lactis exhibits probiotic properties in humans. Considering that Caenorhabditis elegans can be used to study the effects of microorganisms on animal behavior, owing to its simple nervous system, we assessed the impacts of two strains of Lactococcus lactis subsp. Lactis-a non-nisin-producing strain, NBRC 100933 (LL100933), and a nisin-producing strain, NBRC 12007 (LL12007)-on the lifespan, locomotion, reproductive capacity of, and lipid accumulation in, C. elegans. The lifespan of adult C. elegans fed a mixture (1:1) of Escherichia coli OP50 and LL100933 or LL12007 did not show a significant increase compared to that of the group fed a standard diet of E. coli OP50. However, the nematodes fed Lactococcus strains showed notable enhancement in their locomotion at all of the tested ages. Further, the beneficial effects of LL100933 and LL12007 were observed in the daf-16 mutants, but not in the skn-1 and pmk-1 mutants. The lipid accumulation in the worms of the Lactococcus-fed group was lower than that in the control group at all experimental ages. Overall, LL100933 and LL12007 enhance the locomotor behavior of C. elegans, likely by modulating the PMK-1/p38 MAPK and SKN-1/Nrf2 transcription factors.
Keywords: Caenorhabditis elegans; Lactococcus; locomotion; nisin.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
-
- Li T.T., Tian W.L., Gu C.T. Elevation of Lactococcus lactis subsp. cremoris to the Species Level as Lactococcus cremoris sp. nov. and Transfer of Lactococcus lactis subsp. tructae to Lactococcus cremoris as Lactococcus cremoris subsp. tructae comb. nov. Int. J. Syst. Evol. Microbiol. 2019;71:004727. doi: 10.1099/ijsem.0.004727. - DOI - PubMed
-
- Belo G.A., Cordeiro B.F., Oliveira E.R., Braga M.P., Da Silva S.H., Costa B.G., Martins F.D.S., Jan G., Le Loir Y., Gala-García A., et al. SlpB Protein Enhances the Probiotic Potential of L. Lactis NCDO 2118 in Colitis Mice Model. Front. Pharmacol. 2021;12:755825. doi: 10.3389/fphar.2021.755825. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

