WWFedCBMIR: World-Wide Federated Content-Based Medical Image Retrieval
- PMID: 37892874
- PMCID: PMC10604333
- DOI: 10.3390/bioengineering10101144
WWFedCBMIR: World-Wide Federated Content-Based Medical Image Retrieval
Abstract
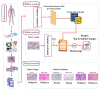
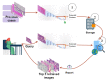
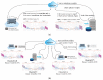
The paper proposes a federated content-based medical image retrieval (FedCBMIR) tool that utilizes federated learning (FL) to address the challenges of acquiring a diverse medical data set for training CBMIR models. CBMIR is a tool to find the most similar cases in the data set to assist pathologists. Training such a tool necessitates a pool of whole-slide images (WSIs) to train the feature extractor (FE) to extract an optimal embedding vector. The strict regulations surrounding data sharing in hospitals makes it difficult to collect a rich data set. FedCBMIR distributes an unsupervised FE to collaborative centers for training without sharing the data set, resulting in shorter training times and higher performance. FedCBMIR was evaluated by mimicking two experiments, including two clients with two different breast cancer data sets, namely BreaKHis and Camelyon17 (CAM17), and four clients with the BreaKHis data set at four different magnifications. FedCBMIR increases the F1 score (F1S) of each client from 96% to 98.1% in CAM17 and from 95% to 98.4% in BreaKHis, with 11.44 fewer hours in training time. FedCBMIR provides 98%, 96%, 94%, and 97% F1S in the BreaKHis experiment with a generalized model and accomplishes this in 25.53 fewer hours of training.
Keywords: breast cancer; computer-aided diagnosis; content-based medical image retrieval (CBMIR); convolutional auto-encoder (CAE); digital pathology; federated learning (FL); histopathological images; whole-slide images (WSIs).
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Figures






References
-
- World Health Organization . Laboratory Quality Standards and Their Implementation. World Health Organization; Geneva, Switzerland: 2011.
Grants and funding
LinkOut - more resources
Full Text Sources

