Identifying Potent Nonsense-Mediated mRNA Decay Inhibitors with a Novel Screening System
- PMID: 37893174
- PMCID: PMC10604367
- DOI: 10.3390/biomedicines11102801
Identifying Potent Nonsense-Mediated mRNA Decay Inhibitors with a Novel Screening System
Abstract
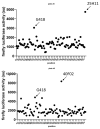
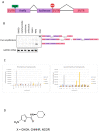
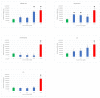
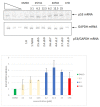
Nonsense-mediated mRNA decay (NMD) is a quality control mechanism that degrades mRNAs carrying a premature termination codon. Its inhibition, alone or in combination with other approaches, could be exploited to develop therapies for genetic diseases caused by a nonsense mutation. This, however, requires molecules capable of inhibiting NMD effectively without inducing toxicity. We have built a new screening system and used it to identify and validate two new molecules that can inhibit NMD at least as effectively as cycloheximide, a reference NMD inhibitor molecule. These new NMD inhibitors show no cellular toxicity at tested concentrations and have a working concentration between 6.2 and 12.5 µM. We have further validated this NMD-inhibiting property in a physiopathological model of lung cancer in which the TP53 gene carries a nonsense mutation. These new molecules may potentially be of interest in the development of therapies for genetic diseases caused by a nonsense mutation.
Keywords: genetic disease; nonsense mutation; nonsense-mediated mRNA decay; small molecules; therapy.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

