Multi-Omic, Histopathologic, and Clinicopathologic Effects of Once-Weekly Oral Rapamycin in a Naturally Occurring Feline Model of Hypertrophic Cardiomyopathy: A Pilot Study
- PMID: 37893908
- PMCID: PMC10603660
- DOI: 10.3390/ani13203184
Multi-Omic, Histopathologic, and Clinicopathologic Effects of Once-Weekly Oral Rapamycin in a Naturally Occurring Feline Model of Hypertrophic Cardiomyopathy: A Pilot Study
Abstract
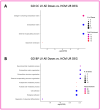
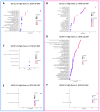
Hypertrophic cardiomyopathy (HCM) remains the single most common cardiomyopathy in cats, with a staggering prevalence as high as 15%. To date, little to no direct therapeutical intervention for HCM exists for veterinary patients. A previous study aimed to evaluate the effects of delayed-release (DR) rapamycin dosing in a client-owned population of subclinical, non-obstructive, HCM-affected cats and reported that the drug was well tolerated and resulted in beneficial LV remodeling. However, the precise effects of rapamycin in the hypertrophied myocardium remain unknown. Using a feline research colony with naturally occurring hereditary HCM (n = 9), we embarked on the first-ever pilot study to examine the tissue-, urine-, and plasma-level proteomic and tissue-level transcriptomic effects of an intermittent low dose (0.15 mg/kg) and high dose (0.30 mg/kg) of DR oral rapamycin once weekly. Rapamycin remained safe and well tolerated in cats receiving both doses for eight weeks. Following repeated weekly dosing, transcriptomic differences between the low- and high-dose groups support dose-responsive suppressive effects on myocardial hypertrophy and stimulatory effects on autophagy. Differences in the myocardial proteome between treated and control cats suggest potential anti-coagulant/-thrombotic, cellular remodeling, and metabolic effects of the drug. The results of this study closely recapitulate what is observed in the human literature, and the use of rapamycin in the clinical setting as the first therapeutic agent with disease-modifying effects on HCM remains promising. The results of this study establish the need for future validation efforts that investigate the fine-scale relationship between rapamycin treatment and the most compelling gene expression and protein abundance differences reported here.
Keywords: autophagy; cat; large animal model; mTOR (mammalian/mechanistic target of rapamycin); proteomics (LCMS); sirolimus; transcriptomics (RNA sequencing); translational.
Conflict of interest statement
S.A.K., S.F., A.F. and L.G. are employees of TriviumVet (Waterford, Ireland), which served as the sponsor of this research. J.A.S. is a member of the scientific advisory for TriviumVet.
Figures






References
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

