Detection of SARS-CoV-2 Δ426 ORF8 Deletion Mutant Cluster in NGS Screening
- PMID: 37894036
- PMCID: PMC10609088
- DOI: 10.3390/microorganisms11102378
Detection of SARS-CoV-2 Δ426 ORF8 Deletion Mutant Cluster in NGS Screening
Abstract
Next-generation sequencing (NGS) from SARS-CoV-2-positive swabs collected during the last months of 2022 revealed a large deletion spanning ORF7b and ORF8 (426 nt) in six patients infected with the BA.5.1 Omicron variant. This extensive genome loss removed a large part of these two genes, maintaining in frame the first 22 aminoacids of ORF7b and the last three aminoacids of ORF8. Interestingly, the deleted region was flanked by two small repeats, which were likely involved in the formation of a hairpin structure. Similar rearrangements, comparable in size and location to the deletion, were also identified in 15 sequences in the NCBI database. In this group, seven out of 15 cases from the USA and Switzerland presented both the BA.5.1 variant and the same 426 nucleotides deletion. It is noteworthy that three out of six cases were detected in patients with immunodeficiency, and it is conceivable that this clinical condition could promote the replication and selection of these mutations.
Keywords: NGS; SARS-CoV-2; deletion; genomic surveillance; variants.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript.
Figures
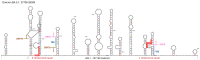



References
-
- Korber B., Fischer W.M., Gnanakaran S., Yoon H., Theiler J., Abfalterer W., Hengartner N., Giorgi E.E., Bhattacharya T., Foley B., et al. Tracking Changes in SARS-CoV-2 Spike: Evidence That D614G Increases Infectivity of the COVID-19 Virus. Cell. 2020;182:812–827.e19. doi: 10.1016/j.cell.2020.06.043. - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

