The Signal Transduction Protein PII Controls the Levels of the Cyanobacterial Protein PipX
- PMID: 37894037
- PMCID: PMC10609283
- DOI: 10.3390/microorganisms11102379
The Signal Transduction Protein PII Controls the Levels of the Cyanobacterial Protein PipX
Abstract
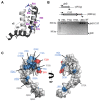
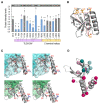
Cyanobacteria, microorganisms performing oxygenic photosynthesis, must adapt their metabolic processes to environmental challenges such as day and night changes. PipX, a unique regulatory protein from cyanobacteria, provides a mechanistic link between the signalling protein PII, a widely conserved (in bacteria and plants) transducer of carbon/nitrogen/energy richness, and the transcriptional regulator NtcA, which controls a large regulon involved in nitrogen assimilation. PipX is also involved in translational regulation through interaction with the ribosome-assembly GTPase EngA. However, increases in the PipX/PII ratio are toxic, presumably due to the abnormally increased binding of PipX to other partner(s). Here, we present mutational and structural analyses of reported PipX-PII and PipX-NtcA complexes, leading to the identification of single amino acid changes that decrease or abolish PipX toxicity. Notably, 4 out of 11 mutations decreasing toxicity did not decrease PipX levels, suggesting that the targeted residues (F12, D23, L36, and R54) provide toxicity determinants. In addition, one of those four mutations (D23A) argued against the over-activation of NtcA as the cause of PipX toxicity. Most mutations at residues contacting PII decreased PipX levels, indicating that PipX stability would depend on its ability to bind to PII, a conclusion supported by the light-induced decrease of PipX levels in Synechococcus elongatus PCC7942 (hereafter S. elongatus).
Keywords: NtcA; PipX toxicity; Synechococcus elongatus; energy sensing; light and dark conditions; mutational analysis; nitrogen regulation network; protein interaction.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
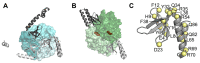

References
-
- Lee H.-W., Noh J.-H., Choi D.-H., Yun M., Bhavya P.S., Kang J.-J., Lee J.-H., Kim K.-W., Jang H.-K., Lee S.-H. Picocyanobacterial Contribution to the Total Primary Production in the Northwestern Pacific Ocean. Water. 2021;13:1610. doi: 10.3390/w13111610. - DOI
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

