A Comparative Study of Growth Performance, Blood Biochemistry, Rumen Fermentation, and Ruminal and Fecal Bacterial Structure between Yaks and Cattle Raised under High Concentrate Feeding Conditions
- PMID: 37894057
- PMCID: PMC10609059
- DOI: 10.3390/microorganisms11102399
A Comparative Study of Growth Performance, Blood Biochemistry, Rumen Fermentation, and Ruminal and Fecal Bacterial Structure between Yaks and Cattle Raised under High Concentrate Feeding Conditions
Abstract
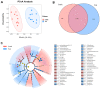
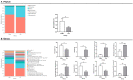
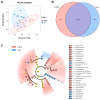
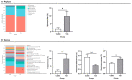
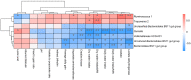
This study compared the growth performance, serum biochemical indicators, rumen fermentation parameters, rumen bacterial structure, and fecal bacterial structure of cattle and yaks fed for two months and given a feed containing concentrate of a roughage ratio of 7:3 on a dry matter basis. Compared with cattle, yak showed better growth performance. The serum biochemical results showed that the albumin/globulin ratio in yak serum was significantly higher than that in cattle. Aspartate aminotransferase, indirect bilirubin, creatine kinase, lactate dehydrogenase, and total cholesterol were significantly lower in yaks than in cattle. The rumen pH, acetate to propionate ratio, and acetate were lower in yaks than in cattle, whereas the lactate in yaks was higher than in cattle. There were significant differences in the structure of ruminal as well as fecal bacteria between cattle and yaks. The prediction of rumen bacterial function showed that there was a metabolic difference between cattle and yaks. In general, the metabolic pathway of cattle was mainly riched in a de novo synthesis of nucleotides, whereas that of yaks was mainly riched in the metabolic utilization of nutrients. This study provides a basis for understanding a rumen ecology under the condition of a high concentrate diet.
Keywords: blood metabolism; cattle; fecal bacterial; rumen bacterial; yaks.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



Similar articles
-
Dietary Conversion from All-Concentrate to All-Roughage Alters Rumen Bacterial Community Composition and Function in Yak, Cattle-Yak, Tibetan Yellow Cattle and Yellow Cattle.Animals (Basel). 2024 Oct 11;14(20):2933. doi: 10.3390/ani14202933. Animals (Basel). 2024. PMID: 39457862 Free PMC article.
-
Comparative study of growth performance, nutrient digestibility, and ruminal and fecal bacterial community between yaks and cattle-yaks raised by stall-feeding.AMB Express. 2021 Jun 30;11(1):98. doi: 10.1186/s13568-021-01259-9. AMB Express. 2021. PMID: 34191139 Free PMC article.
-
Effects of the higher concentrate ratio on the production performance, ruminal fermentation, and morphological structure in male cattle-yaks.Vet Med Sci. 2022 Mar;8(2):771-780. doi: 10.1002/vms3.678. Epub 2021 Dec 17. Vet Med Sci. 2022. PMID: 34918881 Free PMC article.
-
Comparison of rumen bacterial communities between yaks (Bos grunniens) and Qaidam cattle (Bos taurus) fed a low protein diet with different energy levels.Front Microbiol. 2022 Sep 6;13:982338. doi: 10.3389/fmicb.2022.982338. eCollection 2022. Front Microbiol. 2022. PMID: 36147854 Free PMC article.
-
Concentrate supplementation improves cold-season environmental fitness of grazing yaks: responsive changes in the rumen microbiota and metabolome.Front Microbiol. 2023 Aug 28;14:1247251. doi: 10.3389/fmicb.2023.1247251. eCollection 2023. Front Microbiol. 2023. PMID: 37700865 Free PMC article.
Cited by
-
In Vitro Evaluation of Chito-Oligosaccharides on Disappearance Rate of Nutrients, Rumen Fermentation Parameters, and Micro-Flora of Beef Cattle.Animals (Basel). 2024 May 31;14(11):1657. doi: 10.3390/ani14111657. Animals (Basel). 2024. PMID: 38891704 Free PMC article.
-
Effects of High-Concentrate Diets on Growth Performance, Serum Biochemical Indexes, and Rumen Microbiota in House-Fed Yaks.Animals (Basel). 2024 Dec 12;14(24):3594. doi: 10.3390/ani14243594. Animals (Basel). 2024. PMID: 39765498 Free PMC article.
-
Enhancing Feed Efficiency and Growth in Early-Fattening Hanwoo Steers Through High-Energy Concentrate Feeding.Animals (Basel). 2025 Feb 9;15(4):490. doi: 10.3390/ani15040490. Animals (Basel). 2025. PMID: 40002972 Free PMC article.
-
Effects of different dietary energy levels on growth performance, meat quality and nutritional composition, rumen fermentation parameters, and rumen microbiota of fattening Angus steers.Front Microbiol. 2024 May 6;15:1378073. doi: 10.3389/fmicb.2024.1378073. eCollection 2024. Front Microbiol. 2024. PMID: 38770021 Free PMC article.
References
-
- Wei Y.Q., Long R.J., Yang H., Yang H.J., Shen X.H., Shi R.F., Wang Z.Y., Du J.G., Qi X.J., Ye Q.H. Fiber degradation potential of natural co-cultures of Neocallimastix frontalis and Methanobrevibacter ruminantium isolated from yaks (Bos grunniens) grazing on the Qinghai Tibetan Plateau. Anaerobe. 2016;39:158–164. doi: 10.1016/j.anaerobe.2016.03.005. - DOI - PubMed
-
- Liu H., Ran T., Zhang C., Yang W., Wu X., Degen A., Long R., Shi Z., Zhou J. Comparison of rumen bacterial communities between yaks (Bos grunniens) and Qaidam cattle (Bos taurus) fed a low protein diet with different energy levels. Front. Microbiol. 2022;13:982338. doi: 10.3389/fmicb.2022.982338. - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources

