Uncovering Microbiome Adaptations in a Full-Scale Biogas Plant: Insights from MAG-Centric Metagenomics and Metaproteomics
- PMID: 37894070
- PMCID: PMC10608942
- DOI: 10.3390/microorganisms11102412
Uncovering Microbiome Adaptations in a Full-Scale Biogas Plant: Insights from MAG-Centric Metagenomics and Metaproteomics
Abstract
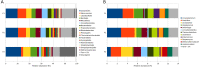
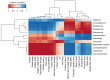
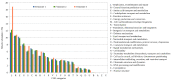
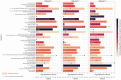
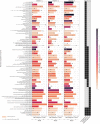
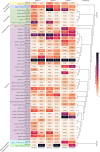
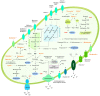
The current focus on renewable energy in global policy highlights the importance of methane production from biomass through anaerobic digestion (AD). To improve biomass digestion while ensuring overall process stability, microbiome-based management strategies become more important. In this study, metagenomes and metaproteomes were used for metagenomically assembled genome (MAG)-centric analyses to investigate a full-scale biogas plant consisting of three differentially operated digesters. Microbial communities were analyzed regarding their taxonomic composition, functional potential, as well as functions expressed on the proteome level. Different abundances of genes and enzymes related to the biogas process could be mostly attributed to different process parameters. Individual MAGs exhibiting different abundances in the digesters were studied in detail, and their roles in the hydrolysis, acidogenesis and acetogenesis steps of anaerobic digestion could be assigned. Methanoculleus thermohydrogenotrophicum was an active hydrogenotrophic methanogen in all three digesters, whereas Methanothermobacter wolfeii was more prevalent at higher process temperatures. Further analysis focused on MAGs, which were abundant in all digesters, indicating their potential to ensure biogas process stability. The most prevalent MAG belonged to the class Limnochordia; this MAG was ubiquitous in all three digesters and exhibited activity in numerous pathways related to different steps of AD.
Keywords: anaerobic digestion; biogas microbiome; biogas process chain; metagenome analyses; metagenomic binning; metaproteome analyses.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
Figures


References
-
- Theuerl S., Klang J., Prochnow A. Process disturbances in agricultural biogas production—Causes, mechanisms and effects on the biogas microbiome: A review. Energies. 2019;12:365. doi: 10.3390/en12030365. - DOI
-
- Hassa J., Maus I., Off S., Pühler A., Scherer P., Klocke M., Schlüter A. Metagenome, metatranscriptome, and metaproteome approaches unraveled compositions and functional relationships of microbial communities residing in biogas plants. Appl. Microbiol. Biotechnol. 2018;102:5045–5063. doi: 10.1007/s00253-018-8976-7. - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Research Materials

